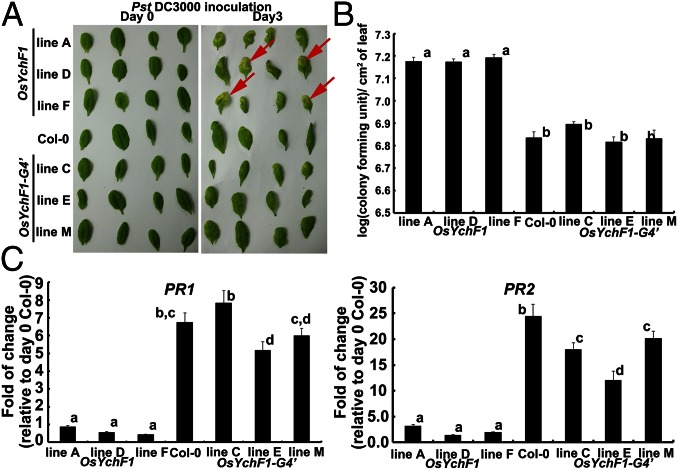
Fig. 4.
The ectopic expression of OsYchF1 but not OsYchF1-G4′ elevated disease symptoms caused by a bacterial pathogen (Pst DC3000). (A) Disease symptoms were observed 3 d after the inoculation of Pst DC3000 via syringe infiltration. More lesions (indicated by red arrows) were observed on the leaves of transgenic Arabidopsis lines expressing OsYchF1, whereas the extents of lesions observed in OsYchF1-G4′ transgenic lines were similar to the untransformed wild-type Col-0. (B) Pathogen titers were determined by counting colony forming units per cm2 of leaf surface area 3 d after inoculation. Error bars: SDs. n = 3 plants. All three transgenic lines expressing OsYchF1 had significantly higher pathogen titers than those expressing OsYchF1-G4′ or Col-0 (wild type). Values associated with the same letters (a or b) were not significantly different from one another (one-way ANOVA, P < 0.05). (C) The expressions of defense marker genes (PR1 and PR2) were estimated by real-time PCR. Error bar: SD. n = at least 3 samples. The transgenic lines expressing OsYchF1 did not show significant levels of induction of PR1 and PR2 after Pst DC3000 inoculation, compared with the wild-type Col-0 at day 0, whereas those expressing OsYchF1-G4′ showed high levels of PR1 and PR2 induction similar to Col-0. All experiments were performed twice, and similar results were obtained.