Abstract
Twenty-three phage-displayed peptides that specifically bind to an anti-benzothiostrobin monoclonal antibody (mAb) in the absence or presence of benzothiostrobin were isolated from a cyclic 8-residue peptide phage library. Competitive and noncompetitive phage enzyme linked immunosorbent assays (ELISAs) for benzothiostrobin were developed by using a clone C3-3 specific to the benzothiostrobin-free mAb and a clone N6-18 specific to the benzothiostrobin immunocomplex, respectively. Under the optimal conditions, the half maximal inhibition concentration (IC50) of the competitive phage ELISA and the concentration of analyte producing 50% saturation of the signal (SC50) of the noncompetitive phage ELISA for benzothiostrobin were 0.94 and 2.27 ng mL−1, respectively. The noncompetitive phage ELISA showed higher selectivity compared to the competitive. Recoveries of the competitive and the noncompetitive phage ELISAs for benzothiostrobin in cucumber, tomato, pear and rice samples were 67.6–119.6% and 70.4–125.0%, respectively. The amounts of benzothiostrobin in the containing incurred residues samples detected by the two types of phage ELISAs were significantly correlated with that detected by high-performance liquid chromatography (HPLC).
Keywords: Benzothiostrobin, Enzyme-linked immunosorbent assay, Phage-displayed peptide, Phage anti-immunocomplex assay, Noncompetitive immunoassay, Peptidomimetics
Graphical Abstract
1. Introduction
Immunoassays are practical analytical tools for detecting pesticides because of their well-known advantages of simplicity, low cost and large parallel-processing capacity [1, 2]. Pesticides are low-molecular-weight chemicals that are buried in the antibody binding pocket after binding, so they are usually detected using a competitive format. In this format, a competitor (a competing hapten conjugated with a carrier protein, enzyme, fluorophore, etc.) that competes with the analyte for binding to the antibody is a necessary reagent [3]. Generally, the sensitivity of the immunoassay can be significantly improved by using heterologous competing haptens, but the chemical synthesis of a series of competing haptens depending on the analyte may be difficult, expensive and potentially hazardous [4, 5]. Besides, some immunoassays have cross-reactivity with analogues of the target because of the analogues can also bind to the antibody [6, 7]. These limitations hinder the widespread use and application of immunoassay in detection of pesticide residues.
The phage display technique offers attractive materials to develop alternate immunoassays that may enhance assay performance by increasing sensitivity and selectivity [8]. Phage-displayed peptide libraries have been a powerful tool for isolating peptides that specifically bind to analyte-free antibodies (peptidomimetics) or analyte-bound antibodies (anti-immunocomplex peptides) [9]. Analyte peptidomimetics can be used as surrogate competing haptens to accelerate the development of heterologous immunoassays. Some investigators have successfully obtained analyte peptidomimetics that can be used as a competing hapten for low-molecular-weight compounds from phage display peptide libraries, and the analyte peptidomimetics show better sensitivity than chemical synthesis of homologous and heterologous competing haptens [3, 9–18]. Anti-immunocomplex phage peptides can specifically recognize an analyte-bound antibody to enhance the affinity and selectivity of the primary antibody because of the formation of a ternary complex, which translates into an improved noncompetitive immunoassay for a low-molecular-weight compound [8]. Although only eight low-molecular-weight compounds (brominated diphenyl ether 47, clomazone, malachite green, leucomalachite green, gibberellin, 2,2′,4,4′-tetrabromodiphenyl ether, phenoxybenzoic acid, molinate and atrazine) have been successfully detected by noncompetitive immunoassays based on anti-immunocomplex phage peptides, each presented better performance than the corresponding conventional competitive immunoassays [8, 9, 19–24]. Additionally, real-time polymerase chain reaction and loop-mediated isothermal amplification have been successfully used to detect low-molecular-weight chemicals by using phage-displayed peptides because of the unique characteristic that phage-displayed peptides can be connected to nucleic acids (single stranded DNA) [3, 25, 26].
Benzothiostrobin, a fungicide developed by the Central China Normal University, is a novel strobilurin fungicide that exhibits highly efficient control of most fungal diseases in crops [27, 28], and patent applications for preparation and use of benzothiostrobin have submitted in China, the United States, Europe, and worldwide (Pub. No.: CN 102302012 B, CN 101379967 A, CN 101268780 B, US 2010/0292285 A1, WO 2007/073637 A1, WO 2009/135407 A1). In our previous study, a homologous competitive enzyme-linked immunosorbent assay (ELISA) was developed for the detection of benzothiostrobin. The half maximal inhibition concentration (IC50) and the limit of detection (LOD, IC10) were 7.55 and 0.428 ng mL−1, respectively. The assay had cross-reactivity (CR) of 0.34% with pyraclostrobin [29].
In this study, the phage-displayed peptides that specifically recognize anti-benzothiostrobin monoclonal antibody (mAb) 4E8 in the absence or presence of benzothiostrobin were isolated from a cyclic 8-amino-acid random peptide library. Both competitive and noncompetitive phage ELISAs for benzothiostrobin based on the isolated phage-diplayed peptides were developed and compared with the conventional ELISA reported earlier [29]. The developed phage ELISAs were validated by high-performance liquid chromatography (HPLC) in the analysis of the samples containing incurred residues.
2. Materials and methods
2.1. Reagents
All reagents were of analytical grade unless specified otherwise. Benzothiostrobin (99.02%) was obtained from the Central China Normal University (Wuhan, China). The pesticide standards used for cross-reactivity studies were purchased from Dr Ehrenstorfer GmbH (Germany). Bovine serum albumin (BSA), tetramethylbenzidine (TMB), isopropyl-β-D-thiogalactoside (IPTG), 5-bromo-4-chloro-3-indolyl- β-D-galactoside (Xgal), and polyoxyethylene sorbitan monolaurate (Tween-20) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). Mouse anti-M13 monoclonal antibody-horse radish peroxidase (HRP) conjugate was purchased from GE Healthcare (Piscataway, NJ). Escherichia coli ER2738 was purchased from New England Biolabs (Ipswich, MA). The mAb 4E8 and the cyclic 8-amino-acid random peptide library (the transducing units (TU) and titer of the library were 5.52×109 pfu μg−1 and 3.4×1013 pfu mL−1, respectively) were developed previously [15, 29].
2.2. Biopanning
Three wells of each microtiter plate were coated with purified 4E8 mAb (10 μg mL−1) in 100 μL of phosphate-buffered saline (PBS) by overnight incubation at 4 °C. Nonspecific binding was blocked by incubation with 300 μL of PBS containing 3% bovine serum albumin (BSA) for 1.5 h at 37 °C. To eliminate nonspecific binding of phage to BSA, another plate coated with 100 μL of 3% BSA in PBS was used for preabsorption. The phage library (1011 phage) diluted with PBS was first added to the preabsorption plate and incubated at room temperature for 1 h. Then, the supernatant was transferred to the plate coated with 4E8 mAb and incubated with shaking at room temperature for 1 h. The wells were washed 10 times with PBS containing 0.1% (v/v) Tween 20 (PBST), and 100 μL of benzothiostrobin (10 μg mL−1 in PBS) was added to each well with shaking for 1 h to compete the binding phage from the coating antibody (peptidomimetics panning). The phage library that had been used for peptidomimetic panning was transferred into other wells of the mAb-coated plate that had been preincubated with 10 μg mL−1 benzothiostrobin and washed five times with PBST, followed by incubation at room temperature for 1 h. The bound phage was eluted with 100 μL of 0.1 mol L−1 glycine-HCl (pH 2.2) per well for 15 min and neutralized with 13 μL of 1 mol L−1 Tris-HCl (pH 9.1) (anti-immunocomplex phage peptides panning). The elution solutions were then collected and used to infect E. coli ER2738 for amplification and titration. The amplified phages were used for a subsequent round of panning. In the second and third rounds of panning, the concentration of coating antibody was reduced to 5 and 2.5 μg mL−1, while the concentration of benzothiostrobin was reduced to 1 and 0.1 μg mL−1, respectively.
After three rounds of panning, 180 μL of ER2738 cell culture (mid-log phase, OD600 = 0.5 AU) was mixed with 10 μL of diluted phage eluates. The infected cells were transferred to culture tubes containing 5 mL 45 °C top agar and poured on a LB/IPTG/Xgal plate. The plates were incubated overnight at 37 °C. A total of 48 clones for peptidomimetics or anti-immunocomplex phage peptides were picked, transferred to diluted ER2738 culture and grown at 37 °C with shaking for 4.5 h. Cells were pelleted by centrifugation at 10000 rpm for 10 min and the supernatants were collected for phage ELISAs. The positive clones as described above were further amplified and used for phage DNA isolation as introduced in the plasmid mini kit instruction manual (Omega Bio-Tek, Inc., GA, USA). The product of phage DNA was submitted for DNA sequencing using the primer 96gIII (CCCTCATAGTTAGCGTAACG) (GenScript Nanjing Co. Ltd., Jiangsu, China).
2.3. Phage ELISAs
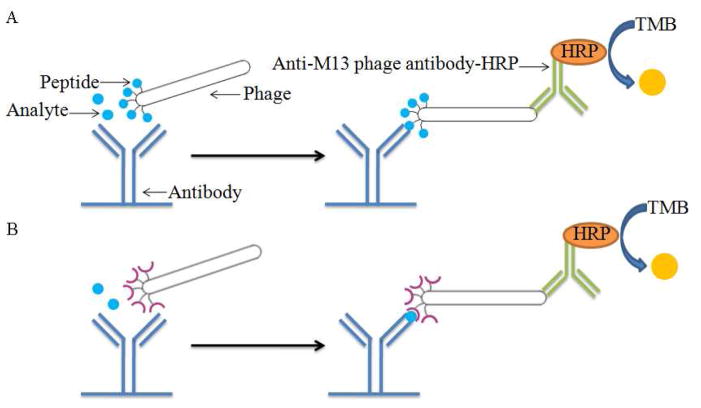
Competitive and noncompetitive phage ELISAs were set up to screen phage capable of binding the benzothiostrobin-free mAb and benzothiostrobin immunocomplex. The microtiter plates were coated with 4E8 mAb at a concentration of 10 μg mL−1 by incubation for 2 h at 37 °C, and blocked for 1.5 h at 37 °C with 1% BSA in PBS. Fifty microliters of phage supernatant of each clone was mixed with 50 μL of 0.2 μg mL−1 benzothiostrobin in 10% methanol-PBS or 10% methanol-PBS. The mixtures were added to the wells and incubated at room temperature for 1 h. After the wells were washed six times with 0.1% PBST, 100 μL of anti-M13 phage antibody conjugated with HRP (1:5000 dilution in PBS) was added. After 1 h incubation and washing six times, the amount of bound enzyme was determined by adding 100 μL of peroxidase substrate (25 mL of 0.1 M citrate acetate buffer (pH 5.5), 0.4 mL of 6 mg mL−1 TMB in dimethyl sulfoxide (DMSO), and 0.1 mL of 1% H2O2). The absorbance at 450 nm was determined after the reaction was stopped by adding 50 μL of 2 M H2SO4 per well (Fig. 1).
Fig. 1.
Schematic diagram of competitive phage ELISA (A) and noncompetitive phage ELISA (B). The 96-well ELISA plate was coated with 1.0 μg well−1 benzothiostrobin antibody. Then 50 μL of phage and 50 μL of PBS with or without benzothiostrobin were added to the wells. A 1:5000 dilution of M13 phage antibody–HRP was used for the detection of bound phage.
2.4. Optimization of the competitive and noncompetitive phage ELISAs
The optimal concentrations of phage and antibody were determined by the checkerboard titration method for the competitive and noncompetitive phage ELISAs [8, 30]. The experimental parameters, including the ionic strength, pH value and organic solvent, were sequentially studied to determine the optimal conditions to achieve the maximum sensitivity. Serial concentrations of benzothiostrobin were detected in PBS solutions containing different concentrations of NaCl (from 0.035 to 3.2 mol L−1) and methanol (from 5 to 40%, v/v) as well as solutions with various pH values (from 4.5 to 9.5) by using the competitive and noncompetitive phage ELISAs. The parameters of the competitive phage ELISA were evaluated using the Amax/IC50 and IC50, and Amax/SC50 and SC50 were used to evaluate the parameters of the noncompetitive phage ELISA.
2.5. Selectivity of the phage ELISA
A series of concentrations (0.01~10 μg mL−1) of the analogues were tested using the competitive and noncompetitive phage ELISAs. For the competitive phage ELISA, the cross-reactivity (CR) was calculated based on the IC50 values using the following formula: CR = [IC50 (benzothiostrobin)/IC50 (analogue)] × 100%. For the noncompetitive phage ELISA, the CR was calculated based on the SC50 values by the formula: CR = [SC50 (benzothiostrobin)/SC50 (analogue)] × 100%. The CR of benzothiostrobin was defined as 100%.
2.6. Analysis of spiked agricultural samples
Four different agricultural samples (cucumber, tomato, pear and rice) were chosen to evaluate the performance of the phage ELISAs. Cucumber, tomato, pear and rice were purchased from local markets. All samples were cut into pieces and homogenized. The homogenized samples were spiked with known concentrations of benzothiostrobin in methanol (the final concentrations were 0.1, 0.2 and 0.3 mg kg−1). The spiked samples were thoroughly mixed and allowed to stand at room temperature overnight.
All samples (10 g) were extracted twice by a vortex mixer in 10 mL of PBS containing 50 % methanol for 1 min and centrifuged for 5 min at 4 000 rpm. The supernatant was transferred into a 25-mL volumetric flask and adjusted to 25 mL with PBS. After appropriate dilution, the solutions were analysed via the competitive and noncompetitive phage ELISAs. Each pesticide was spiked and analysed in triplicate.
2.7. Practical application of the immunoassays
Tomato and rice were collected from farms where benzothiostrobin had been used in Nanjing, China. The concentrations of benzothiostrobin in the samples were simultaneously analyzed by competitive phage ELISA, noncompetitive phage ELISA and HPLC to evaluate the correlation between the methods. For ELISAs, the extraction and analysis of the samples were performed as described above. For HPLC, 10 g of a homogenized tomato or rice sample was weighed in a 250-mLflask, and 10 mL distilled water and 50 mL acetonitrile were added. The sample was vigorously shaken for 1 h at room temperature. The extracts were filtered into a 100 mL cylinder with plug containing 5 g sodium chloride, and the mixture was homogenized and let stand for 30 min. A half of the supernatant was transferred into a 150 mL Florence flask, and concentrated to dryness by a rotatory evaporator at 45°C. The concentrate of tomato was dissolved in 3 mL acetone/n-hexane (5/95, v/v) and loaded onto a Florisil SPE column, that was equilibrated with 5 mL n-hexane. The SPE column was eluted with 5 mL acetone/n-hexane (20/80, v/v). The eluting solution was collected and dried using a vacuum rotary evaporator at 45 °C. The concentrate of rice was dissolved in 10 mL acetone/n-hexane (20/80, v/v) and loaded onto an Alumina-N SPE column, that was equilibrated with 5 mL n-hexane. The effluent liquid was collected and dried using a vacuum rotary evaporator at 45 °C. The residues of tomato and rice were dissolved with 2 mL acetonitrile and detected by HPLC (Agilent 1260) with a SB-C18 column (250 mm×4.6 mm i.d.) using acetonitrile/water (65/35, v/v) as the mobile phase at a flow rate of 0.8 mL min−1 at 30 ºC. The detection wavelength was 230 nm and the injection volume was 20 μL. The concentrations of benzothiostrobin in the analysed samples were calculated by using the calibration curve and regression equation.
3. Results and discussion
3.1. Isolation of phage-displayed peptides
After three rounds of panning, a majority of clones showed significant differences in signal with or without benzothiostrobin in the competitive or noncompetitive phage ELISA (Fig. S1, see electronic supplementary material). Phage DNA from the positive clones was isolated, and the nucleotide sequence of each was determined. Twenty different sequences (designated as C2-3, C3-3, C4-1, C5-2, C7-1, C8-1, C10-1, C12-1, C13-1, C16-1, C17-1, C18-1, C23-1, C24-1, C25-1, C27-1, C36-1, C39-1, C40-1, C45-2) were identified binding to benzothiostrobin-free mAb (Table 1). The clones C2-3 and C18-1 shared the consensus motif of TPXGSL, C3-3 and C23-1 had the consensus sequence GLAXFM, while PXGAWXH was common to C17-1 and C24-1. No evident consensus motif was observed among the other clones. Three different sequences (designated as N1-17, N2-4 and N6-18) were detected binding to the benzothiostrobin immunocomplex (Table 2). The clones N1-17 and N6-18 shared the consensus motif of PXIWPXXW, where X represents different amino acids. These results indicated that the motifs of TPXGSL, GLAXFM and PXGAWXH contributed significantly to binding to the benzothiostrobin-free mAb, and the motif of PXIWPXXW contributed significantly to the benzothiostrobin immunocomplex recognition.
Table 1.
Peptide sequences isolated with the benzothiostrobin-free mAb.
| Clone name | Sequencea | Clone name | Sequence |
|---|---|---|---|
| C2-3 | PATPLGSL (3) | C39-1 | PWYVPQGS (1) |
| C18-1 | KGTPMGSL (1) | C4-1 | GTPYGSLK (1) |
| C3-3 | SGLAEFMS (3) | C5-2 | EGPLRSIN (2) |
| C23-1 | TGLAPFMK (1) | C8-1 | LTHADLDY (1) |
| C7-1 | LAGADFHV (1) | C10-1 | QTAFGMLP (1) |
| C17-1 | PIGAWYHI (1) | C12-1 | IYHEGHSM (2) |
| C24-1 | PQGAWHHL (1) | C16-1 | PNTWIAHA (1) |
| C13-1 | PSTYLPGA (1) | C25-1 | LPQHLLAS (1) |
| C27-3 | PWYYLPGF (3) | C36-1 | MLGPRDNE (1) |
| C45-2 | PWPWATPL (2) | C40-1 | IPNMMGRS (1) |
A total of 29 clones were sequenced. The numbers of isolates bearing the same sequence are indicated in parentheses.
Table 2.
Peptide sequences isolated with the benzothiostrobin immune complex.
| Clone name | Sequencea |
|---|---|
| N1-17 | PNIWPESW (17) |
| N2-4 | LHSKHTYE (4) |
| N6-18 | PDIWPTAW (18) |
A total of 39 clones were sequenced. The numbers of isolates bearing the same sequence are indicated in parentheses.
3.2. Selection of phage-displayed peptide
The positive clones that specifically bind to mAb 4B8 were employed to develop competitive assays for benzothiostrobin. The concentrations of phage and antibody are shown in Table S1, and the IC50 values of benzothiostrobin were in the range 0.83–6.28 ng mL−1 (Table S2). The clone C3-3 was superior to the other phage-displayed peptides in terms of sensitivity. Each positive clone that was specifically for the benzothiostrobin immunocomplex was used in a noncompetitive assay for benzothiostrobin. Serial dilutions of phage particles were added to the wells of the plate coated with four different concentrations of mAb 4B8 (10, 5, 2.5, and 1.25 μg mL−1) in the absence or presence of benzothiostrobin (0 or 100 ng mL−1). The maximal signal difference was observed at antibody concentration of 10 μg mL−1 and phage concentrations of 1.25×109 pfu mL−1 for N1-17 and N2-4, and 7.8×107 pfu mL−1 for N6-18 (Fig. S2 to S4). The assay setup with clone N6-18 performed with the highest sensitivity (SC50= 3.40 ng mL−1), followed by clones N1-17 and N2-4 (SC50values of 5.77 and 3.44 ng mL−1, respectively) (Fig. S5).
3.3. Optimization of the phage ELISAs
Competitive and noncompetitive phage ELISAs are based on antigen-antibody interactions, and the ionic strength and pH value are additional factors influencing the equilibrium constant [31]. Therefore, the sensitivities of the assays are often improved by optimizing the ionic strength and pH value of the working solution. The pH 7.4, 0.14 M NaCl PBS buffer was selected for the competitive phage ELISA because it exhibited the highest Amax/IC50 and lower IC50 (Table S3 and S4). When the PBS buffer contained 0.2 mol L−1 NaCl at pH 7, the noncompetitive phage ELISA exhibited the highest Amax/SC50 and lower SC50 (Fig. S6 and S7).
Organic solvent can improve the solubility of the analytical, but high concentration of organic solvent often affects antigen-antibody binding. In addition, organic solvents are generally used to extract analyte from the samples, and the sample extract is directly diluted with buffer for removing the matrix interference. Therefore, the effect of organic solvent must be studied in the optimization of an immunoassay. Methanol is commonly selected as an organic co-solvent in immunoassays because of its weaker effect on antigen-antibody binding. When the final concentration of methanol was less than or equal to 2.5% (50 μL of phage supernatant in PBS was mixed with 50 μL of analyte in 5% methanol-PBS), the effects of methanol on the phage ELISAs were negligible (Fig. S8 and S9). In conclusion, the optimum parameters were 2.5% methanol and 0.14 M NaCl at pH 7.4 for the competitive phage ELISA, 2.5% methanol and 0.2 M NaCl at pH 7 for the noncompetitive phage ELISA.
3.4. Sensitivity of the phage ELISAs
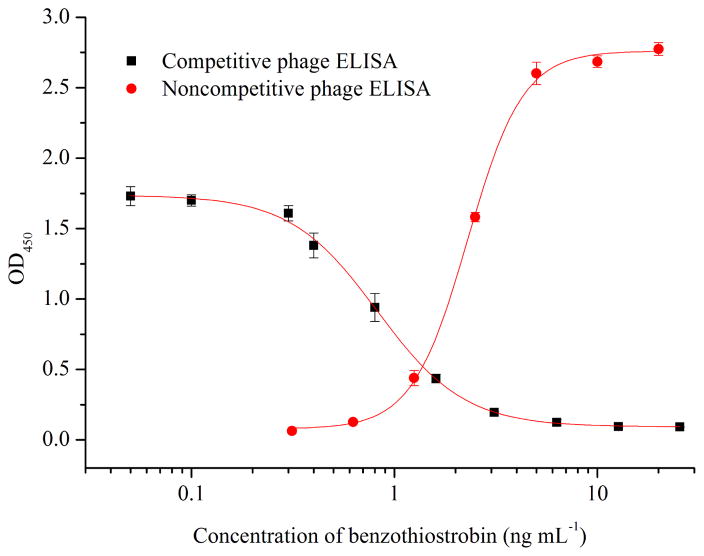
Under the optimum conditions, the standard curves of benzothiostrobin were obtained using the relationship between the OD450 value and the concentration of benzothiostrobin shown in Fig. 2. The IC50 values, IC10 values and liner range (IC10 to IC90) of the competitive phage ELISA were 0.94 0.22 and 0.22 to 3.94 ng mL−1, while the SC50 values, SC10 values and liner range (SC10 to SC90) of the noncompetitive phage ELISA were 2.27, 1.11 and 1.11 to 4.62 ng mL−1, respectively. The competitive phage ELISA showed higher sensitivity and wider liner range compared to the noncompetitive. However, the liner range and sensitivity were tunable by adjusting the amount of phage peptide or anti-M13 phage antibody–HRP [8]. In our previous study, a conventional competitive ELISA with an IC50 of 7.55 ng mL−1 and IC10 of 0.43 ng mL−1 was developed by using the same mAb 4B8 [29]. As compared with the IC50 value of the conventional competitive ELISA, the sensitivity of the competitive and noncompetitive phage ELISAs were improved more than 8-fold and 3-fold, respectively. For the HPLC, a good linearity was acquired for benzothiostrobin within the range of 50–10000 ng mL−1, and the equation of the linear regression was y=120.0x-0.702 (R2= 1). The LOD (signal-to-noise (S/N) ratio of 3) and limit of quantification (LOQ, S/N ratio of 10) of the HPLC were 1.30 and 4.33 ng mL−1, respectively. Therefore, the phage and conventional ELISAs showed higher sensitivity compared to the HPLC.
Fig. 2.
Standard curves for benzothiostrobin by competitive and noncompetitive phage ELISAs. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptides in optimized buffer, then 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
3.5. Selectivity of the phage ELISAs
Table 3 shows the CR results of the competitive and noncompetitive phage ELISA for the analogues of benzothiostrobin. The noncompetitive phage ELISA exhibited no CR (less than 0.03%) for the analogues when the concentration of tested compounds was 10 μg mL−1. The competitive phage ELISA showed 0.34% CR for pyraclostrobin, which conformed to the conventional competitive ELISA in our previous study [29]. In this study, the phage-displayed peptide N6-18 could specifically recognize the benzothiostrobin immunocomplex to form a ternary complex which promoted an increased selectivity for the analyte. Therefore, the noncompetitive format showed higher selectivity than the competitive format. This is consistent with previously reported results [22].
Table 3.
IC50and SC50 values and cross-reactivity of a set of analogues structurally related to benzothiostrobin by competitive and noncompetitive phage ELISAs.
| Compound | Chemical structure | Competitive | Noncompetitive | ||
|---|---|---|---|---|---|
|
| |||||
| IC50 (ng mL−1) | CR (%) | SC50 (ng mL−1) | CR (%) | ||
| Benzothiostrobin |
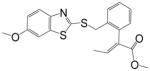
|
0.94 | 100 | 2.27 | 100 |
| Pyraclostrobin |
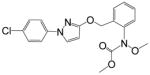
|
275.3 | 0.34 | >10000 | <0.03 |
| Azoxystrobin |
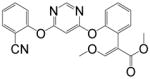
|
>10000 | <0.01 | >10000 | <0.03 |
| Kresoxim-methyl |
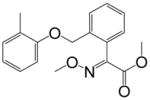
|
>10000 | <0.01 | >10000 | <0.03 |
| Picoxystrobin |
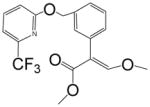
|
>10000 | <0.01 | >10000 | <0.03 |
| Chloropiperidine Ester |
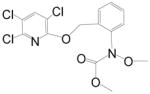
|
>10000 | <0.01 | >10000 | <0.03 |
| Fluoxastrobin |
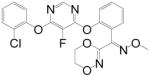
|
>10000 | <0.01 | >10000 | <0.03 |
| Fenaminstrobin |
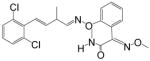
|
>10000 | <0.01 | >10000 | <0.03 |
3.6. Analysis of spiked samples
Due to the excellent sensitivity and selectivity of an immunoassay, matrix interference can be simply eliminated by diluting with buffer. When the extracts of the cucumber, tomato, pear and rice were diluted equal or more than 8-fold (the total dilution was equal or more than 40-fold, containing 2-fold by mixing with phage in the phage ELISA procedure and 2.5-fold during extraction), the standard curves of the competitive and noncompetitive immunoassays prepared during the matrix dilution were similar to those in the matrix-free buffer (Fig. S10 and S11). Therefore, the spiked samples were diluted at least 40-fold to remove the matrix interferences before the phage ELISA analysis. As shown in Table 4, the mean recoveries and the relative standard deviations (RSDs) were 67.6–119.6% and 2.2–13.7% for the competitive phage ELISA, and 70.4–125.0% and 1.8–11.5 for the noncompetitive phage ELISA, respectively.
Table 4.
Average recoveries of samples spiked with benzothiostrobin by competitive and noncompetitive phage ELISAs (n=3).
| Sample | Spiked (ng g−1) | Competitive | Noncompetitive | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Measured±SD (ng g−1) | Average recovery (%) | RSD (%) | Measured±SD (ng g−1) | Average recovery (%) | RSD (%) | ||
| Cucumber | 100 | 71.0 ±9.1 | 71.0 | 12.8 | 118.6±9.6 | 118.6 | 8.1 |
| 200 | 200.1 ±5.6 | 100.0 | 2.8 | 221.6±11.6 | 110.8 | 5.2 | |
| 300 | 206.2 ±4.5 | 68.7 | 2.2 | 232.7±26.8 | 77.6 | 11.5 | |
| Tomato | 100 | 98.0 ±6.4 | 98.0 | 6.6 | 115.7±12.5 | 115.7 | 10.8 |
| 200 | 239.3 ±18.4 | 119.6 | 7.7 | 235.1±4.2 | 117.5 | 1.8 | |
| 300 | 339.9 ±11.2 | 113.3 | 3.3 | 240.0±12.5 | 80.0 | 5.2 | |
| Pear | 100 | 74.1 ±5.9 | 74.1 | 8.0 | 125.0±5.5 | 125.0 | 4.4 |
| 200 | 237.3 ±12.4 | 118.7 | 5.2 | 217.2±14.1 | 108.6 | 6.5 | |
| 300 | 307.0 ±12.3 | 102.3 | 3.9 | 211.3±9.7 | 70.4 | 4.6 | |
| Rice | 100 | 67.6 ±9.3 | 67.6 | 13.7 | 89.5±7.7 | 89.5 | 8.6 |
| 200 | 187.6 ±21.7 | 93.8 | 11.6 | 193.5±13.9 | 96.7 | 7.2 | |
| 300 | 341.7 ±13.9 | 113.9 | 4.1 | 225.4±8.7 | 75.1 | 3.9 | |
Fig. S12 shows the representative HPLC chromatograms of the standard benzothiostrobin sample, blank sample and spiked sample. These chromatograms indicated that the matrix interferences of tomato and rice were removed by the sample treatments. The accuracy and precision of the HPLC method was evaluated by measuring the spiked tomato and rice samples (the final concentrations were 100, 200 and 300 ng g−1), the average recoveries ranged from 80.8% to 100.5%, and the RSDs were less than or equal to 5.5% (Table S5).
3.7. Analysis of samples containing incurred residues
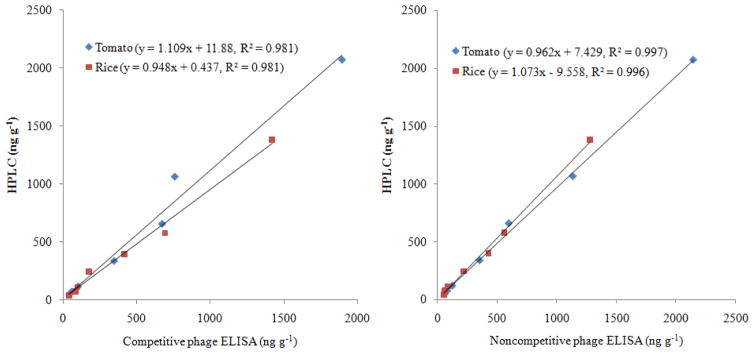
Table 5 shows the results from the immunoassays and HPLC for benzothiostrobin-positive tomato and rice samples in Nanjing, China. The concentrations of benzothiostrobin in tomato samples as determined using the competitive phage ELISA, noncompetitive phage ELISA and HPLC were in the ranges 43.1–1890 ng g−1, 61.4–2150 ng g−1 and 50.3–2070 ng g−1, respectively. The concentrations in rice determined using the same methods as above were in the ranges 38.9–1420 ng g−1, 53.8–1280 ng g−1 and 41.1–1380 ng g−1, respectively. The P values were greater than 0.05, implied that the data between the HPLC and phage ELISAs were not significantly different. Besides, the phage ELISAs and HPLC techniques yielded comparable results and good correlations (Fig. 3). Therefore, the competitive and noncompetitive phage ELISAs presented in this study were reliable and accurate.
Table 5.
Comparison of benzothiostrobin residues between the phage ELISAs and HPLC in authentic samples.
| Matrix | Method | Sample (ng g−1)
|
P valuea | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| Tomato | HPLC | 120 | 339 | 659 | 2070 | 76.0 | 50.3 | 1070 | - |
| Competitive | 102 | 347 | 673 | 1890 | 59.9 | 43.1 | 758 | 0.172 | |
| Noncompetitive | 124 | 353 | 598 | 2140 | 81.3 | 61.4 | 1130 | 0.398 | |
| Rice | HPLC | 246 | 41.1 | 398 | 74.9 | 1380 | 578 | 114 | - |
| Competitive | 174 | 38.9 | 415 | 84.2 | 1420 | 692 | 96.8 | 0.575 | |
| Noncompetitive | 219 | 53.8 | 430 | 62.1 | 1280 | 562 | 88.1 | 0.258 | |
Significant difference assessment between the HPLC and the phage ELISAs by using Student’s t test.
Fig. 3.
Correlations between the phage ELISAs and HPLC for the samples containing incurred residues. Fourteen tomato and rice samples obtained from fields treated with benzothiostrobin were detected by competitive phage ELISA, noncompetitive phage ELISA and HPLC. The equation of the line and correlation coefficient obtained from linear regression of the combined phage ELISAs and HPLC data for benzothiostrobin in tomato or rice samples is shown.
4. Conclusions
In the current study, twenty-three phage-displayed peptides were isolated from the cyclic 8-residue peptide phage library for simultaneous development of competitive and noncompetitive format immunoassays. The IC50 values of the competitive phage ELISA and the SC50 values of the noncompetitive phage ELISA were 0.94 and 2.27 ng mL−1 respectively, which were improved more than 8-fold and 3-fold compared to the conventional competitive ELISA with an IC50 of 7.55 ng mL−1. The competitive phage ELISA presented higher sensitivity and wider liner range compared to the noncompetitive. The noncompetitive format showed higher selectivity than both the phage-peptide and conventional competitive formats. The two formats of phage ELISAs present similar accuracies and precisions in the analysis of agricultural samples. The accuracy and precision of the assays were validated by HPLC, and results were comparable with correlations > R2=0.98. Therefore, the phage ELISAs presented in this study are suitable as convenient quantitative tools for the rapid screening of benzothiostrobin in agricultural products. These results demonstrate that the presented phage-displayed peptide technology is a convenient and efficient method for the developments of heterologous competitive and noncompetitive format immunoassays for small molecules to enhance the assay performance.
Supplementary Material
Fig. S1. Screening of positive clones by competitive and noncompetitive phage ELISAs. A: twenty-nine clones out of 48 showed significant signal differences with or without benzothiostrobin by competitive phage ELISA; B: thirty-nine clones out of 48 showed significant signal differences with or without benzothiostrobin by noncompetitive phage ELISA.
Fig. S2. Reactivity of the phage-displayed peptide N1–17 with the benzothiostrobin immunocomplex using different amounts of coating antibody. Plates were coated with antibody at 10 μg mL−1 (A), 5 μg mL−1 (B), 2.5 μg mL−1 (C), and 1.25 μg mL−1 (D) and were incubated with serial dilutions of phage-displayed peptide in the presence or absence of 100 ng mL−1 benzothiostrobin.
Fig. S3. Reactivity of the phage-displayed peptide N2–4 with the benzothiostrobin immunocomplex using different amounts of coating antibody. Plates were coated with antibody at 10 μg mL−1 (A), 5 μg mL−1 (B), 2.5 μg mL−1 (C), and 1.25 μg mL−1 (D) and were incubated with serial dilutions of phage-displayed peptide in the presence or absence of 100 ng mL−1 benzothiostrobin.
Fig. S4. Reactivity of the phage-displayed peptide N6–18 with the benzothiostrobin immunocomplex using different amounts of coating antibody. Plates were coated with antibody at 10 μg mL−1 (A), 5 μg mL−1 (B), 2.5 μg mL−1 (C), and 1.25 μg mL−1 (D) and were incubated with serial dilutions of phage-displayed peptide in the presence or absence of 100 ng mL−1 benzothiostrobin.
Fig. S5. Dose–response curves for phage clones N1–17, N2–4, N6–18. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptides in 5% methanol-PBS. Next 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
Fig. S6. Effect of pH value on noncompetitive phage ELISA. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptide in 5% methanol-PBS with different pH values, and 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
Fig. S7. Effect of ionic strength on noncompetitive phage ELISA. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptide in 5% methanol-PBS containing different ionic strengths, and 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
Fig. S8. Effect of methanol on competitive phage ELISA. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptide in PBS containing different concentrations of methanol, and 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
Fig. S9. Effect of methanol on noncompetitive phage ELISA. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptide in PBS containing different concentrations of methanol, and 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
Fig. S10. Matrix interference on competitive phage ELISA. Standard inhibition curves for benzothiostrobin in the buffer, cucumber (A), tomato (B), pear (C) and rice (D) matrices using the competitive phage ELISA.
Fig. S11. Matrix interference on noncompetitive phage ELISA. Standard binding curves for benzothiostrobin in the buffer, cucumber (A), tomato (B), pear (C) and rice (D) matrices using the noncompetitive phage ELISA.
Fig. S12. The representative chromatograms of HPLC. Standard benzothiostrobin sample (A), blank sample of tomato (B), spiked sample of tomato (C), positive sample of tomato (D), blank sample of rice (E), spiked sample of rice (C), positive sample of rice (D).
Table S1 The optimal concentrations of phage and antibody for competitive phage ELISA.
Table S2 Average IC50 and Amax/IC50 values of the twenty competitive phage ELISAs.
Table S3 Average IC50 and Amax/IC50 values of the competitive phage ELISA in PBS solutions of various pH.
Table S4 Average IC50 and Amax/IC50 values of the competitive phage ELISA in PBS solutions containing different concentrations of NaCl.
Table S5 Recoveries of samples spiked with benzothiostrobin by HPLC.
Highlights.
Benzothiostrobin peptidomimetics have been isolated.
Anti-benzothiostrobin immunocomplex phage-displayed peptides have been obtained.
Competitive and noncompetitive phage ELISAs have been developed.
The IC50 and SC50 of the phage ELISAs are 0.94 and 2.27 ng mL−1, respectively.
The phage ELISAs have better performance than conventional ELISA.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31431794), the National Natural Science Foundation of China (31301690) and the Doctoral Program of Higher Education Research Fund (20130097120006). Research supported in part by grants from the National Institute of Environmental Health Science Superfund Research Program (P42 ES04699) and the Centers for Disease Control, National Institute of Occupational Health Science (2U50 OH007550).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morozova VS, Levashova AI, Eremin SA. Determination of pesticides by enzyme immunoassay. J Anal Chem. 2005;60:202–217. [Google Scholar]
- 2.Farré M, Kantiani L, Barceló D. Advances in immunochemical technologies for analysis of organic pollutants in the environment. Trends Anal Chem. 2007;26:1100–1112. [Google Scholar]
- 3.Kim HJ, González-Techera A, González-Sapienza GG, Ahn KC, Gee SJ, Hammock BD. Phage-borne peptidomimetics accelerate the development of polyclonal antibody-based heterologous immunoassays for the detection of pesticide metabolites. Environ Sci Technol. 2008;42:2047–2053. doi: 10.1021/es702219a. [DOI] [PubMed] [Google Scholar]
- 4.Lee WY, Lee EK, Kim YJ, Park WC, Chung T, Lee YT. Monoclonal antibody-based enzyme-linked immunosorbent assays for the detection of the organophosphorus insecticide isofenphos. Anal Chim Acta. 2006;557:169–178. [Google Scholar]
- 5.Gui WJ, Liu YH, Wang CM, Liang X, Zhu GN. Development of a direct competitive enzyme-linked immunosorbent assay for parathion residue in food samples. Anal Biochem. 2009;393:88–94. doi: 10.1016/j.ab.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Kolosova AY, Park JH, Eremin SA, Park SJ, Kang SJ, Shim WB, Lee HS, Lee YT, Chung DH. Comparative study of three immunoassays based on monoclonal antibodies for detection of the pesticide parathion-methyl in real samples. Anal Chim Acta. 2004;511:323–331. [Google Scholar]
- 7.Sun JW, Dong TT, Zhang Y, Wang S. Development of enzyme linked immunoassay for the simultaneous detection of carbaryl and metolcarb in different agricultural products. Anal Chim Acta. 2010;666:76–82. doi: 10.1016/j.aca.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 8.Kim HJ, Rossotti MA, Ahn KC, González-Sapienza GG, Gee SJ, Musker R, Hammock BD. Development of a noncompetitive phage anti-immunocomplex assay for brominated diphenyl ether 47. Anal Biochem. 2010;401:38–46. doi: 10.1016/j.ab.2010.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Liu ZP, Li GQ, Li J, Kim HJ, Shelver WL, Li QX, Xu T. Simultaneous development of both competitive and noncompetitive immunoassays for 2,2′,4,4′-tetrabromodiphenyl ether using phage-displayed peptides. Anal Bioanal Chem. 2013;405:9579–9583. doi: 10.1007/s00216-013-7364-5. [DOI] [PubMed] [Google Scholar]
- 10.Cardozo S, González-Techera A, Last JA, Hammock BD, Kramer K, González-Sapienza GG. Analyte peptidomimetics selected from phage display peptide libraries: a systematic strategy for the development of environmental immunoassays. Environ Sci Technol. 2005;39:4234–4241. doi: 10.1021/es047931l. [DOI] [PubMed] [Google Scholar]
- 11.Thirumala-Devi K, Miller JS, Reddy G, Reddy DVR, Mayo MA. Phage-displayed peptides that mimic aflatoxin B1 in serological reactivity. J Appl Microbiol. 2001;90:330–336. doi: 10.1046/j.1365-2672.2001.01249.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu RR, Yu Z, He QH, Xu Y. An immunoassay for ochratoxin A without the mycotoxin. Food Control. 2007;18:872–877. [Google Scholar]
- 13.Hua XD, Liu XF, Shi HY, Wang YR, Kim HJ, Gee SJ, Wang MH, Liu FQ, Hammock BD. Development of a heterologous enzyme-linked immunosorbent assay for organophosphorus pesticides with phage-borne peptide. RSC Adv. 2014;4:42445–42453. doi: 10.1039/C4RA07059C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González-Techera A, Umpiérrez-Failache M, Cardozo S, Obal G, Pritsch O, Last JA, Gee SJ, Hammock BD, González-Sapienza G. High-throughput method for ranking the affinity of peptide ligands selected from phage display libraries. Bioconjug Chem. 2008;19:993–1000. doi: 10.1021/bc700279y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YR, Wang H, Li PW, Zhang Q, Kim HJ, Gee SJ, Hammock DB. Phage-displayed peptide that mimics aflatoxins and its application in immunoassay. J Agric Food Chem. 2013;61:2426–2433. doi: 10.1021/jf4004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He QH, Xu Y, Huang YH, Liu RR, Huang ZB, Li YP. Phage-displayed peptides that mimic zearalenone and its application in immunoassay. Food Chem. 2011;126:1312–1315. [Google Scholar]
- 17.Liu RR, Xu L, Qiu XM, Chen XL, Deng SL, Lai WH, Xu Y. An immunoassay for determining aflatoxin B1 using a recombinant phage as a nontoxic coating conjugate. J Food Safety. 2012;32:318–325. [Google Scholar]
- 18.Yuan QP, Pestka JJ, Hespenheide BM, Kuhn LA, Linz JE, Hart LP. Identification of mimotope peptides which bind to the mycotoxin deoxynivalenol-specific monoclonal antibody. Appl Environ Microb. 1999;65:3279–3286. doi: 10.1128/aem.65.8.3279-3286.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González-Techera A, Kim HJ, Gee SJ, Last JA, Hammock BD, González-Sapienza G. Polyclonal antibody-based noncompetitive immunoassay for small analytes developed with short peptide loops isolated from phage libraries. Anal Chem. 2007;79:9191–9196. doi: 10.1021/ac7016713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inaba J, Nakamura S, Shimizu K, Asami T, Suzuki Y. Anti-metatype peptides, a molecular tool with high sensitivity and specificity to monitor small ligands. Anal Biochem. 2009;388:63–70. doi: 10.1016/j.ab.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Dong JX, Xu C, Wang H, Xiao ZL, Gee SJ, Li ZF, Wang F, Wu WJ, Shen YD, Yang JY, Sun YM, Hammock BD. Enhanced sensitive immunoassay: non-competitive phage anti-immune complex assay for the determination of malachite green and leucomalachite green. J Agric Food Chem. 2014;62:8752–8758. doi: 10.1021/jf5019824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossotti MA, Carlomagno M, González-Techera A, Hammock BD, Last J, González-Sapienza G. Phage anti-immunocomplex assay for clomazone: two-site recognition increasing assay specificity and facilitating adaptation into an on-site format. Anal Chem. 2010;82:8838–8843. doi: 10.1021/ac101476f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HJ, Ahn KC, González-Techera A, González-Sapienza GG, Gee SJ, Hammock BD. Magnetic bead-based phage anti-immunocomplex assay (PHAIA) for the detection of the urinary biomarker 3-phenoxybenzoic acid to assess human exposure to pyrethroid insecticides. Anal Biochem. 2009;386:45–52. doi: 10.1016/j.ab.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.González-Techera A, Vanrell L, Last JA, Hammock BD, González-Sapienza G. Phage anti-immune complex assay: general strategy for noncompetitive immunodetection of small molecules. Anal Chem. 2007;79:7799–7806. doi: 10.1021/ac071323h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua XD, Yin W, Shi HY, Li M, Wang YR, Wang H, Ye YH, Kim HJ, Gee SJ, Wang MH, Liu FQ, Hammock BD. Development of phage immuno-loop-mediated isothermal amplification assays for organophosphorus pesticides in agroproducts. Anal Chem. 2014;86:8441–8447. doi: 10.1021/ac5020657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HJ, McCoy M, Gee SJ, González-Sapienza GG, Hammock BD. Noncompetitive phage anti-immunocomplex real-time polymerase chain reaction for sensitive detection of small molecules. Anal Chem. 2011;83:246–253. doi: 10.1021/ac102353z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao PL, Wang F, Huan W, Chen Q, Liu ZM. Synthesis and fungicidal activities of novel thioethers containing benzothiazole moiety. Chin J Org Chem. 2010;30:1567–1573. [Google Scholar]
- 28.Xu CY, Hou YP, Wang JX, Yang GF, Liang XY, Zhou MG. Activity of a novel strobilurin fungicide benzothiostrobin against Sclerotinia sclerotiorum. Pestic Biochem Phys. 2014;115:32–38. doi: 10.1016/j.pestbp.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Yuan YL, Hua XD, Li M, Yin W, Shi HY, Wang MH. Development of a sensitive indirect competitive enzyme-linked immunosorbent assay based on the monoclonal antibody for the detection of benzothiostrobin residue. RSC Adv. 2014;4:24406–24411. [Google Scholar]
- 30.Shan GM, Wengatz I, Stoutamire DW, Gee SJ, Hammock BD. An enzyme-linked immunosorbent assay for the detection of esfenvalerate metabolites in human urine. Chem Res Toxicol. 1999;12:1033–1041. doi: 10.1021/tx990091h. [DOI] [PubMed] [Google Scholar]
- 31.Reverberi R, Reverberi L. Factors affecting the antigen-antibody reaction. Blood Transfus. 2007;5:227–240. doi: 10.2450/2007.0047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Screening of positive clones by competitive and noncompetitive phage ELISAs. A: twenty-nine clones out of 48 showed significant signal differences with or without benzothiostrobin by competitive phage ELISA; B: thirty-nine clones out of 48 showed significant signal differences with or without benzothiostrobin by noncompetitive phage ELISA.
Fig. S2. Reactivity of the phage-displayed peptide N1–17 with the benzothiostrobin immunocomplex using different amounts of coating antibody. Plates were coated with antibody at 10 μg mL−1 (A), 5 μg mL−1 (B), 2.5 μg mL−1 (C), and 1.25 μg mL−1 (D) and were incubated with serial dilutions of phage-displayed peptide in the presence or absence of 100 ng mL−1 benzothiostrobin.
Fig. S3. Reactivity of the phage-displayed peptide N2–4 with the benzothiostrobin immunocomplex using different amounts of coating antibody. Plates were coated with antibody at 10 μg mL−1 (A), 5 μg mL−1 (B), 2.5 μg mL−1 (C), and 1.25 μg mL−1 (D) and were incubated with serial dilutions of phage-displayed peptide in the presence or absence of 100 ng mL−1 benzothiostrobin.
Fig. S4. Reactivity of the phage-displayed peptide N6–18 with the benzothiostrobin immunocomplex using different amounts of coating antibody. Plates were coated with antibody at 10 μg mL−1 (A), 5 μg mL−1 (B), 2.5 μg mL−1 (C), and 1.25 μg mL−1 (D) and were incubated with serial dilutions of phage-displayed peptide in the presence or absence of 100 ng mL−1 benzothiostrobin.
Fig. S5. Dose–response curves for phage clones N1–17, N2–4, N6–18. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptides in 5% methanol-PBS. Next 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
Fig. S6. Effect of pH value on noncompetitive phage ELISA. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptide in 5% methanol-PBS with different pH values, and 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
Fig. S7. Effect of ionic strength on noncompetitive phage ELISA. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptide in 5% methanol-PBS containing different ionic strengths, and 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
Fig. S8. Effect of methanol on competitive phage ELISA. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptide in PBS containing different concentrations of methanol, and 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
Fig. S9. Effect of methanol on noncompetitive phage ELISA. Serial dilutions of benzothiostrobin standard were mixed with phage-displayed peptide in PBS containing different concentrations of methanol, and 100 μL of the mixtures were added to the antibody-coated wells. Each point represents the mean value of three replicates.
Fig. S10. Matrix interference on competitive phage ELISA. Standard inhibition curves for benzothiostrobin in the buffer, cucumber (A), tomato (B), pear (C) and rice (D) matrices using the competitive phage ELISA.
Fig. S11. Matrix interference on noncompetitive phage ELISA. Standard binding curves for benzothiostrobin in the buffer, cucumber (A), tomato (B), pear (C) and rice (D) matrices using the noncompetitive phage ELISA.
Fig. S12. The representative chromatograms of HPLC. Standard benzothiostrobin sample (A), blank sample of tomato (B), spiked sample of tomato (C), positive sample of tomato (D), blank sample of rice (E), spiked sample of rice (C), positive sample of rice (D).
Table S1 The optimal concentrations of phage and antibody for competitive phage ELISA.
Table S2 Average IC50 and Amax/IC50 values of the twenty competitive phage ELISAs.
Table S3 Average IC50 and Amax/IC50 values of the competitive phage ELISA in PBS solutions of various pH.
Table S4 Average IC50 and Amax/IC50 values of the competitive phage ELISA in PBS solutions containing different concentrations of NaCl.
Table S5 Recoveries of samples spiked with benzothiostrobin by HPLC.