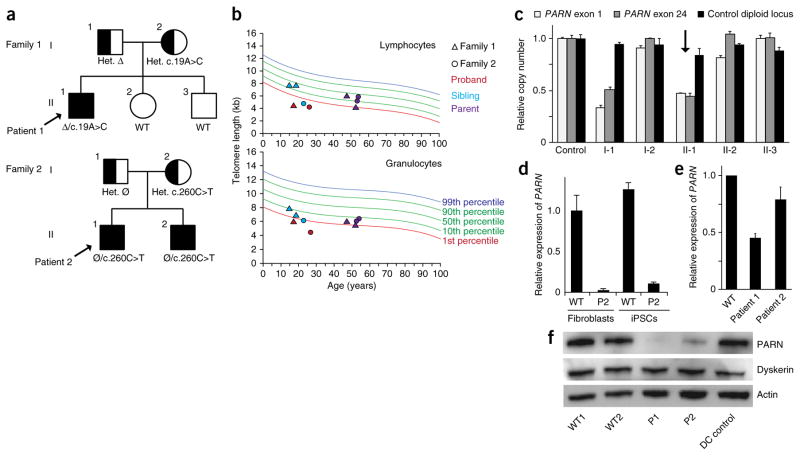
Figure 1.
PARN mutations in two families with dyskeratosis congenita. (a) Segregation of compound heterozygous PARN mutations in families 1 and 2. Probands are indicated by arrows. Clinically affected individuals are shown as filled shapes, and carriers are shown as half-filled shapes. Nucleotide substitutions are indicated, as well as the inheritance of a large deletion (Δ) encompassing the PARN locus in family 1 and a noncoding defect affecting accumulation of PARN transcripts in family 2 (Ø). Het., heterozygous; WT, wild type. (b) Telomere length measurement by flow-FISH in lymphocytes (top) and granulocytes (bottom). (c) Quantitative PCR (qPCR) of relative PARN copy number in peripheral blood cell genomic DNA from a control and members of family 1, using primers from the first (exon 1) and last (exon 24) exons of the PARN locus, as compared to copy number for a control diploid locus (GPR15) (n = 2 technical replicates). Error bars, s.d. The proband is indicated by an arrow. (d) qPCR of cDNA using an allele-specific primer that distinguishes wild-type (c.C260) from mutant (c.T260) transcripts (n = 2 technical replicates). Error bars, s.d. Cells from patient 2 (P2) show deficits in transcript levels from the ‘normal’ allele (lacking a protein-coding mutation), which are expected to be 50% of those in wild-type cells. (e) qPCR of PARN transcripts from patient-derived and normal fibroblasts (n = 3 biological replicates). Error bars, s.d. (f) Immunoblot of PARN, dyskerin and actin protein levels in fibroblasts from normal individuals with wild-type PARN, patients 1 and 2 with PARN mutations, and a patient with dyskeratosis congenita without PARN mutations (DC control).