Abstract
Rhesus cytomegalovirus (RhCMV) 68-1 is the prototypic strain of RhCMV that has been used for pathogenesis and vaccine development. We determined the complete sequence of the RhCMV 68-1 UL/b’ region directly from the original urine from which RhCMV 68-1 was isolated in 1968, and compared it to other RhCMVs. The laboratory passaged RhCMV 68-1 has inversions, deletions, and stop codons in UL/b’ that are absent in the original isolate and other low passage RhCMV isolates. Fourteen of the 17 open reading frames (ORFs) in 68-1 UL/b’ in the original isolate share >95% amino acid identity with low passage RhCMV. The original isolate retains 6 ORFs that encode α-chemokine-like proteins, including RhUL146 and RhUL146b that share only 92% and 81% amino acid identity, respectively, with a contemporary low passage RhCMV isolate. Identification of the original RhCMV 68-1 UL/b’ sequence is important for using RhCMV 68-1 in pathogenesis and vaccine studies.
Keywords: human cytomegalovirus, rhesus cytomegalovirus, Cercopithecine herpesvirus, macacine herpesvirus 3, cynomolgus macaque cytomegalovirus, UL/b’
Introduction
Human cytomegalovirus (HCMV) causes congenital disease in infants and neonates, mononucleosis in adults, and severe disease in transplant recipients and immunocompromised patients (Mocarski et al., 2013). Currently, an effective vaccine for human CMV (HCMV) has not been licensed, although it has been given a priority 1 status by the Institute of Medicine (Stratton et al., 1990). Rhesus cytomegalovirus (RhCMV) infection in rhesus macaques mimics infection of HCMV in humans, making RhCMV the most practical animal model that most closely resembles HCMV (Powers and Fruh, 2008). Like HCMV, most infections with RhCMV are asymptomatic, result from mucosal contact with bodily fluids containing the virus, and develop into a lifelong latent infection. Experimental infection of macaques with RhCMV can cause congenital disease in neonates and disseminated disease in immunocompromised animals (Yue and Barry, 2008). RhCMV, like HCMV, is accompanied by hematologic changes that may include monocytosis, neutropenia, and lymphocytosis. Both RhCMV and HCMV are common in their host populations (Andrade et al., 2003; Bate et al., 2010; Kessler et al., 1989; Vogel et al., 1994). About 80% of RhCMV open reading frames (ORFs) have orthologs in HCMV and ≥90% have orthologs at the level of protein families, strongly suggesting a common ancestor (Hansen et al., 2003; Malouli et al., 2012; Oxford et al., 2008; Rivailler et al., 2006). Comparison of the sequence of RhCMV ORFs with those from other Old World monkey CMV isolates showed a high level of conservation which was validated by identification of virion proteins using mass spectrometry (Malouli et al., 2012). Both RhCMV and HCMV contain a highly labile region labeled UL/b’ that encodes proteins important in cellular tropism and immune evasion. The UL/b’ region of RhCMV and HCMV frequently undergoes mutation during in vitro passage when virus is propagated in fibroblasts (Hansen et al., 2003; Oxford et al., 2008; Revello and Gerna, 2010; Rivailler et al., 2006).
RhCMV was first isolated from the urine of healthy rhesus macaques (Macaca mulatta) in 1968, propagated in fibroblasts, and the resulting strain was termed 68-1 (Asher et al., 1969; Asher et al., 1974). The cytopathic effects observed in vitro were similar to those seen with HCMV and African green monkey CMV strains, with refractile rounding and sloughing of infected human fetal lung fibroblasts. RhCMV 68-1, deposited in the American Type Culture Collection after subsequent passages in primary rhesus fibroblasts, was completely sequenced in 2003 (Hansen et al., 2003). Interestingly, after propagation in fibroblasts, RhCMV 68-1 was reported to replicate poorly in endothelial and epithelial cells, which mimicked the in vitro effects of highly-passaged HCMV laboratory strains like AD169 (Lilja and Shenk, 2008). Subsequently, it was determined that the UL/b’ region of laboratory passaged strains of HCMV and RhCMV (namely 68-1 and 180.92) have large deletions compared with low passage (or unpassaged) isolates (Cha et al., 1996; Dolin et al., 2004; Hansen et al., 2003; Murphy and Shenk, 2003; Rivailler et al., 2006). The UL/b’ region of CMV contains UL128, UL130, and UL131A, which are required for infection of endothelial cells and epithelial cells (Hahn et al., 2004; Lilja and Shenk, 2008; Ryckman et al., 2006; Wang and Shenk, 2005) as well as other viral proteins which are likely important for virus structure (Spaderna et al., 2005), signaling (Cheung et al., 2005), virus spread (Penfold et al., 1999), immune evasion (Wills et al., 2005), and latency (Goodrum et al., 2007). With the exception of replication of virus in endothelial and epithelial cells, the other functions of the UL/b’ region have not been formally shown to be conserved in RhCMV. Only RhCMV laboratory strains that retain a fully intact UL/b’ set of genes are shed at high levels and are easily transmitted from animal to animal in primate colonies (Oxford et al., 2011).
RhCMV68-1 has been used to study RhCMV pathogenesis (Lockridge et al. 1999), as a model for CMV vaccine development (Yue et al. 2007), and most recently as a vector to induce long-term (Hansen et al 2011) and very broad CD8 T cell responses to SIV (Hansen et al. 2013). To better understand the evolution of the prototype strain of RhCMV68-1, we determined the sequence of the UL/b’ region of the virus directly from the original urine obtained in 1968 by Asher et al. and compared it with the sequence of other strains of RhCMV, other primate CMVs, and HCMV. We found that the 68-1 UL/b’ ORF organization and sequence closely aligns with sequences from low passage contemporary RhCMV isolates, and that the inversions and deletions in the laboratory strain of 68-1 are notably absent. Analysis of the original sequence of the UL/b’ region of 68-1 suggests that the regions important for cellular tropism and immune evasion are highly sensitive to in vitro selection, and that the greatest variance occurs amongst the CXC-chemokine-like genes within the region.
Results and Discussion
Sequence of the UL/b’ Region from RhCMV 68-1-P0
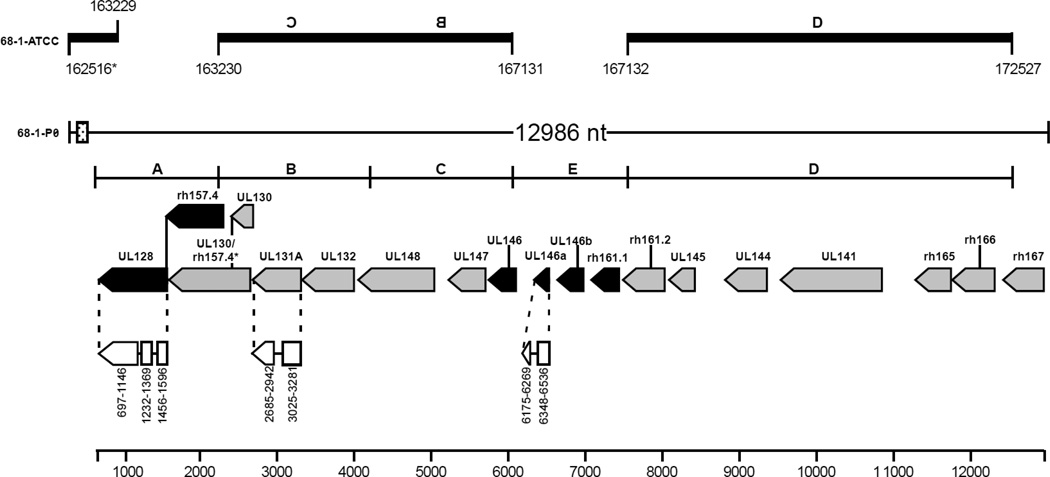
Asher originally obtained urine from 9 juvenile rhesus macaques in 1968 (Asher 1969). Animals were hydrated with intravenous fluid and given mannitol to induce diuresis. Penicillin and streptomycin were added to the urine and a portion was frozen at −70°C. Urine that had not been frozen was serially diluted, and 0.2 ml aliquots of undiluted or diluted urine were used to inoculate stationary WI38 cell (fetal human lung diploid fibroblast) tube cultures. CPE was first noted in cells inoculated with diluted urine from a female macaque on day 17 and virus from undiluted urine from the same animal was passaged in WI38 cell tube cultures and deposited in the ATCC as RhCMV 68-1 (Asher et al., 1974). Despite repeated attempts we were unable to isolate infectious virus from the urine. However, sufficient virus DNA remained in the urine sample to amplify by PCR; the copy number of RhCMV DNA in the urine was 18.3 copies of RhCMV DNA/uL by real time-PCR. We isolated DNA from the urine and obtained overlapping PCR products that corresponded to the UL/b’ region RhCMV 68-1 P0 (passage 0). The complete region of RhCMV 68-1 P0 UL/b’ (GenBank accession #KF011492) is 12.9 kilobase pairs with a GC content of 46% and encodes 17 ORFs in the same orientation which are most conserved with low-passage RhCMV isolates from naturally-infected macaques (Malouli et al., 2012; Oxford et al., 2008) (Figure 1, Table 1). While RhCMV 68-1-ATCC is missing ORFs UL128, UL146a, UL146b, rh161.1 and a portion of UL130, the RhCMV 68-1-P0 strain contains full-length sequences for each of these ORFs. The 5’ end of RhCMV 68-1-P0 UL/b’ has a 22 nucleotide sequence repeated 18 times, 16 of which are direct repeats. These sequences are conserved throughout various strains of simian CMVs, (Supplemental Table 1, nucleotide polymorphisms underlined and bolded) and may encode regulatory miRNAs (Hancock et al., 2012).
Fig. 1.
Genomic arrangement of UL/b’ open reading frames (ORFs) sequenced from the original source of RhCMV 68-1-P0 compared with passaged RhCMV 68-1 deposited in the ATCC (68-1-ATCC). Nucleotide numbering of RhCMV 68-1-ATCC (based on Rivailler et al., 2006) is shown at the top for comparison. The genomic structures, consensus regions (A-D), and predicted ORFs are depicted based on previously reported sequences of RhCMVs from low passage or unpassaged isolates (Rivailler et al., 2006; Oxford et al., 2008). RhCMV 68-1-P0 ORFs are on the complementary strand. Gray shaded ORFs are present or have a highly homologous region in 68-1-ATCC, while black shaded ORFs are absent from 68-1-ATCC. White regions joined by lines are proposed of exons. The splice variant for UL130 is indicated as UL130/rh157.4. A putative microRNA region (22-mer repeated 18 times, dotted box) is highlighted at the 5’ end of the contig and is also present in the 68-1-ATCC strain. The figure is based on that from Oxford et al. 2008.
Table 1.
Amino acid percent maximum identity comparison of RhCMV 68-1-P0 UL/b’ open reading frames to other CMV isolates.
| % AA Maximum Identity of 68-1-P0 vs. Other CMV Isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| RhCMV ORF |
Protein Class | Proposed Function | RhCMV 68-1 (ATCC) |
RhCMV CNPRC |
RhCMV 22659 |
RhCMV 180.92 |
Cyno CMV (Ottawa) |
Chimp CMV |
HCMV (Merlin) |
| UL128 | Membrane glycoprotein |
Pentamer envelope complex Essential for endo/epithelial cell tropism |
NH | 99 | 100 | 99 | 97 | 36 | 421 |
| UL130 | Membrane glycoprotein |
Pentamer envelope complex Essential for endo/epithelial cell tropism |
1002 | 98 | 99 | 99 | 87 | 80 | 80 |
| UL131A | Membrane glycoprotein |
Pentamer envelope complex Essential for endo/epithelial cell tropism |
1002 | 93 | 91 | 95 | 97 | 78 | 60 |
| UL132 (rh160) |
Membrane glycoprotein |
Essential for epithelial cell tropism | 99 | 99 | 99 | 99 | 95 | 35 | 40 |
| UL148 (rh159) |
Potential CXC (α) Chemokine |
Potentially encoded as a large transcript with UL146-UL132 |
100 | 99 | 99 | 1003 | 89 | 55 | 42 |
| UL147 (rh158) |
Potential CXC Chemokine |
Homology to HCMV vCXCL2α-chemokine like protein |
100 | 97 | 99 | NH | 98 | 44 | 41 |
| UL146 | Potential CXC Chemokine |
Homology to HCMV vCXCL1 α-chemokine like protein |
100 | 92 | 100 | NH | 79 | UL146: 35 UL146A: 56 |
26 |
| UL146a | Potential CXC Chemokine |
α-chemokine like protein | NH | 98 | 99 | NH | 90 | 33 | UL146: 57 |
| UL146b | Potential CXC Chemokine |
α-chemokine like protein | NH | 81 | 100 | NH | 80 | NH | NH |
| rh161.1 | Potential CXC Chemokine |
Possible duplication of rh161.2 α-chemokine like protein |
344 | 98 | 100 | NH | 93 | NH | NH |
| rh161.24 | Potential CXC Chemokine |
α-chemokine like protein | 1004 | 98 | 100 | NH | 97 | NH | NH |
| UL145 (rh162) |
Potential kinase | Highly conserved with putative PKC and CK II sites |
100 | 100 | 99 | NH | 99 | 65 | 65 |
| UL144 (rh163) |
Membrane Receptor |
TNFR-like protein | 100 | 97 | 99 | NH | 99 | 43 | 63 |
| UL141 (rh164) |
Membrane glycoprotein |
Downregulate cell surface expression of CD155 and CD112 |
100 | 99 | 99 | NH | 97 | 47 | 67 |
| rh165 | Membrane glycoprotein |
Possible duplication of tandem ORFs (rh165, 166, 167) |
100 | 99 | 96 | NH | 95 | NH | NH |
| rh166 | Membrane glycoprotein |
Possible duplication of tandem ORFs (rh165, 166, 167) |
100 | 99 | 98 | NH | 96 | NH | NH |
| rh167 | Membrane glycoprotein |
Possible duplication of tandem ORFs (rh165, 166, 167) |
100 | 98 | NH | 973 | 95 | NH | NH |
GenBank numbers: RhCMV 68-1-P0 (KF011492); RhCMV 68-1-ATCC (AY186194.1); RhCMV CNPRC (EF990255.1); RhCMV 22659 (EU130540.1); RhCMV 180.92 (DQ120516.1); CynoCMV-Ottawa (JN227533.1); Chimp CMV (AF480884.1); HCMV-Merlin (AY446894.2)
ADB84700.1 (low-passage clinical isolate used due to mutation in UL128 of Merlin.)
Comparison made using nucleotide alignment
ORF truncated in comparison sequence
Comparison made using rh161.2 (which was formerly termed rh161 in 68-1-ATCC [Oxford et al. 2008])
NH=Homolog not found in comparison sequence
Comparison of RhCMV 68-1-P0 with other simian CMV UL/b’ ORFs
The 17 ORFs encoded in RhCMV 68-1 P0 UL/b’ were compared with their orthologs in other simian CMVs based on bit scores to establish if one ORF is more closely related to a certain strain of CMV. Bit scores (calculated using NCBI BLASTp) are normalized log-scale sequence alignment scores that compare sequence alignments taking into account maximum sequence identity (longest stretch of matching amino acids) and total sequence identity (overall number of matching amino acids); the higher the number, the less likely the alignment was to have occurred by pure chance and the more likely the sequences are to have originated from similar sources. The bit scores for RhCMV 68-1-P0 UL/b’genes show that these ORFs are most similar to those in RhCMV CNPRC (not passaged in vitro), 68-1-ATCC, and 22659 (a low passage isolate) (Supplemental Figure 1, data not shown). As reported previously, RhCMV shares greatest homology outside of its species with cynomolgus macaque cytomegalovirus (CynoCMV) and does not have as high of a degree of homology with chimpanzee CMV or HCMV (Table 1).
Amino acid alignment of ORFs truncated or missing from 68-1-ATCC UL/b’
Comparison of ORFs in UL/b’ between RhCMV 68-1-P0 and 68-1-ATCC indicates that ORFs UL128, UL146a, UL146b, and rh161.1 are missing from 68-1-ATCC, while ORF UL130 (which encodes a membrane glycoprotein) is truncated. RhCMV 68-1-P0 encodes UL146, UL146a, UL146b, UL147, rh161.1 and rh161.2, which are putative CXC chemokines (Alcendor et al., 2009; Oxford et al., 2008). These genes in are thought to have arisen from a single ancestral CXCL gene that gave rise to daughter genes and subsequent gene duplications (Alcendor et al., 2009). RhCMV 68-1-ATCC, encodes only UL146, UL147, and rh161.2 (which was formerly termed rh161 [Oxford et al. 2008] in 68-1-ATCC). HCMV UL146 activates CXCR1 and CXCR2, probably to attract neutrophils that carry virus to endothelial cells (Luttichau 2010), while UL147 is also important for neutrophil chemotaxis (Penfold et al., 1999). UL146a is an ELR- CXC chemokine, which suggests that the protein is important for leukocyte migration (Ebert et al., 2005), while UL146b is an ELR+ CXC chemokine, which implies that it can interact with CXCR1 and CXCR2 and induce migration of neutrophils (Kobayashi 2008). rh161.1 and rh161.2 are thought to be the result of a gene duplication event. While most other proteins in UL/b’ share >95% amino acid identity between RhCMV 68-1-P0 and other RhCMV isolates, genes encoding the putative CXC chemokines are the least conserved between different RhCMV isolates. UL146b and UL146 share only 81% and 92% amino acid identity between RhCMV 68-1-P0 and RhCMV CNPRC (Table 1), and rh161.1 in RhCMV 68-1-P0 and rh161.2 from RhCMV 68-1-ATCC share only 34% amino acid identity.
UL128 is a glycoprotein, derived from exon splicing, that is one of the components of the CMV pentameric complex (consisting of UL128, UL130, UL131A, gH and gL) important for epithelial and endothelial cell tropism (Lilja and Shenk 2008; Ryckman et al., 2006). Antibody to the pentameric complex is the predominant component of CMV neutralizing antibody in human sera (Fouts et al., 2012; Genini et al., 2011; Macagno et al. 2010), and immunization of monkeys with the pentameric complex induces broadly neutralizing antibody in the animals (Wussow et al., 2013). The RhCMV 68-1-P0 UL/b’ region contains a full-length copy of UL128, which includes all three exons, and its amino acid sequence is highly conserved among other RhCMV and CynoCMV isolates (Supplemental Figure 2). While the initial description of the CynoCMV Ottawa strain did not report having all three exons (Marsh et al., 2011), analysis of the nucleotide sequence revealed that CynoCMV does have a potential third exon in UL128 (Supplemental Figure 2, gray highlight).
UL130 is also a glycoprotein produced from exon splicing that is part of the pentameric complex. While the spliced version of UL130 from RhCMV 68-1-P0 aligns well with other RhCMV and CynoCMV sequences (Supplemental Figure 3), RhCMV 68-1-P0 has a deletion of 16 amino acids compared with other isolates, which likely is a result of a duplication event. RhCMV 68-1-ATCC only has the first exon of UL130, and is missing the second exon which has been termed rh157.4 (Oxford et al. 2008). Although not reported in the initial publications (Marsh et al., 2011; Rivailler et al., 2006), analysis of both RhCMV 180.92 and CynoCMV nucleotide sequences reveals that each contains the second exon of UL130 (Supplemental Figure 3, gray highlight).
UL131A is another glycoprotein within the pentameric complex. Of the three glycoproteins in this complex encoded by the UL/b’ region, UL131A shares the least homology between RhCMV 68-1-P0 and other RhCMV isolates ranging from 91-95% amino acid identity.
Conclusions
The organization and sequence of the UL/b’ region from the original isolate of RhCMV 68-1 (RhCMV 68-1-P0) closely aligns with contemporary wild-type isolates of RhCMV, and inversions and deletions in the laboratory strain of 68-1-ATCC relative to contemporary isolates are absent. The regions in the UL/b’ region of RhCMV 68-1-P0 important for cellular tropism and immune evasion are highly sensitive to in vitro selection and the greatest divergence occurs in the CXC-chemokine-like genes.
Materials and methods
RhCMV viral DNA was isolated from urine using the QIAamp ® Viral RNA Mini Kit (QIAGEN Inc., Valencia, CA) according to the manufacturer’s instructions. RhCMV genome copy number in the urine was determined by real-time PCR using TaqMan® FAM(6-carboxyfluorescein)/TAMRA(tetramethylrhodamine) technology (Applied Biosystems, Foster City, CA), using primers 5’-TGCGTACTATGGAAGAGACAATGC (gB RT Forward) and 5’-ACATCTGGCCGTTCAAAAAAAC (gB RT Reverse) and a FAM-tagged probe 5’-FAM/ CCAGAAGTTGCGCATCCGCTT to determine RhCMV gB copy number.
The UL/b’ region of RhCMV 68-1 was amplified by PCR (primers listed in Supplemental Table 2). Overlapping PCR products obtained by using the HotStarTaq® DNA polymerase kit (QIAGEN Inc.) were gel purified (GenEluteTM Agarose Spin Column, Sigma-Aldrich, St. Louis, MO). Amplicons were cloned using the Promega pGEM®-T Easy Vector System (Madison, WI) and sequenced using either external T7 and SP6 or internal primers. At least two clones for each amplicon were sequenced. If clones from amplicons were not obtained, the isolated fragments were sequenced directly. Sequencing reactions were assembled using Lasergene SeqMan Pro (DNASTAR, Inc., Madison, WI). The UL/b’ sequence from RhCMV 68-1-P0 has been deposited in Genbank (accession number KF011492).
Supplementary Material
Acknowledgments
This study was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases. We thank Anita Mora and Heather Murphy of Rocky Mountain Laboratories (NIAID) for their assistance with artwork, Yanmei Wang for assistance with viral DNA quantification, and Peter Barry for advice with sequencing.
References
- Alcendor DJ, Zong J, Dolan A, Gatherer D, Davison AJ, Hayward GS. Patterns of divergence in the vCXCL and vGPCR gene clusters in primate cytomegalovirus genomes. Virology. 2009;395:21–32. doi: 10.1016/j.virol.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MR, Yee J, Barry P, Spinner A, Roberts JA, Cabello PH, Leite JP, Lerche NW. Prevalence of antibodies to selected viruses in a long-term closed breeding colony of rhesus macaques (Macaca mulatta) in Brazil. Am J Primatol. 2003;59:123–128. doi: 10.1002/ajp.10069. [DOI] [PubMed] [Google Scholar]
- Asher DM, Gibbs CJ, Jr, Lang DJ. Rhesus monkey cytomegalovirus: persistent asymptomatic viruria (abstract V269) Bacteriol Proc. 1969;69:191. [Google Scholar]
- Asher DM, Gibbs CJ, Jr, Lang DJ, Gajdusek DC. Persistent shedding of cytomegalovirus in the urine of healthy rhesus monkeys. Proc Soc Exp Biol Med. 1974;145:794–801. doi: 10.3181/00379727-145-37897. [DOI] [PubMed] [Google Scholar]
- Bate SL, Dollard SC, Cannon MJ. Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988–2004. Clin Infect Dis. 2010;50:1439–1447. doi: 10.1086/652438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TC, Humphreys IR, Potter KG, Norris PS, Shumway HM, Tran BR, Patterson G, Jean-Jacques R, Yoon M, Spear PG, Murphy KM, Lurain NS, Benedict CA, Ware CF. Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc Natl Acad Sci U S A. 2005;102:13218–13223. doi: 10.1073/pnas.0506172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Holton M, Dolan A, Dargan DJ, Gatherer D, Hayward GS. Comparative genomics of primate cytomegaloviruses. In: Reddehase MJ, editor. Cytomegaloviruses: from Molecular Pathogenesis to Intervention. Norwich, UK: Caister Academic Press; 2013. pp. 1–22. [Google Scholar]
- Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. Genetic content of wild-type human cytomegalovirus. J Gen Virol. 2004;85:1301–1312. doi: 10.1099/vir.0.79888-0. [DOI] [PubMed] [Google Scholar]
- Ebert LM, Schaerli P, Moser B. Chemokine-mediated control of T cell traffic in lymphoid and peripheral tissues. Mol Immunol. 2005;42:799–809. doi: 10.1016/j.molimm.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Fouts AE, Chan P, Stephan JP, Vandlen R, Feierbach B. Antibodies against the gH/gL/UL128/UL130/UL131 complex comprise the majority of the anti-CMV neutralizing antibody response in CMV-HIG. J. Virol. 2012;86:7444–7447. doi: 10.1128/JVI.00467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Früh K, Malouli D, Oxford KL, Barry PA. Non-human-primate models of cytomegalovirus infection, prevention, and therapy. In: Reddehase MJ, editor. Cytomegaloviruses: from Molecular Pathogenesis to Intervention. Norwich, UK: Caister Academic Press; 2013. pp. 463–496. [Google Scholar]
- Genini E, Percivalle E, Sarasini A, Revello MG, Baldanti F, Gerna G. Serum antibody response to the gH/gL/pUL128-131 five-protein complex of human cytomegalovirus (HCMV) in primary and reactivated HCMV infections. J Clin Virol. 2011;52:113–118. doi: 10.1016/j.jcv.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Goodrum F, Reeves M, Sinclair J, High K, Shenk T. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood. 2007;110:937–945. doi: 10.1182/blood-2007-01-070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol. 2004;78:10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock MH, Tirabassi RS, Nelson JA. Rhesus cytomegalovirus encodes seventeen microRNAs that are differentially expressed in vitro and in vivo. Virology. 2012;425:133–142. doi: 10.1016/j.virol.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M, Jr, Lifson JD, Picker LJ. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SG, Sacha JB, Hughes CM, Ford JC, Burwitz BJ, Scholz I, Gilbride RM, Lewis MS, Gilliam AN, Ventura AB, Malouli D, Xu G, Richards R, Whizin N, Reed JS, Hammond KB, Fischer M, Turner JM, Legasse AW, Axthelm MK, Edlefsen PT, Nelson JA, Lifson JD, Früh K, Picker LJ. Cytomegalovirus Vectors Violate CD8+ T Cell Epitope Recognition Paradigms. Science. 2013;340:1237874. doi: 10.1126/science.1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol. 2003;77:6620–6636. doi: 10.1128/JVI.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MJ, London WT, Madden DL, Dambrosia JM, Hilliard JK, Soike KF, Rawlins RG. Serological survey for viral diseases in the Cayo Santiago rhesus macaque population. P R Health Sci J. 1989;8:95–97. [PubMed] [Google Scholar]
- Kobayashi Y. The role of chemokines in neutrophil biology. Front Biosci. 2008;13:2400–2407. doi: 10.2741/2853. [DOI] [PubMed] [Google Scholar]
- Lilja AE, Shenk T. Efficient replication of rhesus cytomegalovirus variants in multiple rhesus and human cell types. Proc Natl Acad Sci U S A. 2008;105:19950–19955. doi: 10.1073/pnas.0811063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockridge KM, Sequar G, Zhou SS, Yue Y, Mandell CP, Barry PA. Pathogenesis of experimental rhesus cytomegalovirus infection. J Virol. 1999;73:9576–9583. doi: 10.1128/jvi.73.11.9576-9583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüttichau HR. The cytomegalovirus UL146 gene product vCXCL1 targets both CXCR1 and CXCR2 as an agonist. J Biol Chem. 2010;285:9137–9146. doi: 10.1074/jbc.M109.002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macagno A, Bernasconi NL, Vanzetta F, Dander E, Sarasini A, Revello MG, Gerna G, Sallusto F, Lanzavecchia A. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol. 2010;84:1005–1013. doi: 10.1128/JVI.01809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malouli D, Nakayasu ES, Viswanathan K, Camp DG, 2nd, Chang WL, Barry PA, Smith RD, Fruh K. Reevaluation of the coding potential and proteomic analysis of the BAC-derived rhesus cytomegalovirus strain 68-1. J Virol. 2012;86:8959–8973. doi: 10.1128/JVI.01132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh AK, Willer DO, Ambagala AP, Dzamba M, Chan JK, Pilon R, Fournier J, Sandstrom P, Brudno M, MacDonald KS. Genomic sequencing and characterization of cynomolgus macaque cytomegalovirus. J Virol. 2011;85:12995–13009. doi: 10.1128/JVI.05840-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ES, Shenk T, Griffiths P, Pass RF. Cytomegalovirus. In: Knipe DM, Howley PM, Cohen JI, Griffith DE, Lamb RA, Martin MA, Racaniello V, Roizman B, editors. Fields Virology. 6th. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1960–2014. [Google Scholar]
- Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci USA. 2003;100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford KL, Eberhardt MK, Yang KW, Strelow L, Kelly S, Zhou SS, Barry PA. Protein coding content of the UL)b' region of wild-type rhesus cytomegalovirus. Virology. 2008;373:181–188. doi: 10.1016/j.virol.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford KL, Strelow L, Yue Y, Chang WL, Schmidt KA, Diamond DJ, Barry PA. Open reading frames carried on UL/b' are implicated in shedding and horizontal transmission of rhesus cytomegalovirus in rhesus monkeys. J Virol. 2011;85:5105–5114. doi: 10.1128/JVI.02631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold ME, Dairaghi DJ, Duke GM, Saederup N, Mocarski ES, Kemble GW, Schall TJ. Cytomegalovirus encodes a potent alpha chemokine. Proc Natl Acad Sci U S A. 1999;96:9839–9844. doi: 10.1073/pnas.96.17.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers C, Fruh K. Rhesus CMV: an emerging animal model for human CMV. Med Microbiol Immunol. 2008;197:109–115. doi: 10.1007/s00430-007-0073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revello MG, Gerna G. Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications. Rev Med Virol. 2010;20:136–155. doi: 10.1002/rmv.645. [DOI] [PubMed] [Google Scholar]
- Rivailler P, Kaur A, Johnson RP, Wang F. Genomic sequence of rhesus cytomegalovirus 180.92: insights into the coding potential of rhesus cytomegalovirus. J Virol. 2006;80:4179–4182. doi: 10.1128/JVI.80.8.4179-4182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol. 2006;80:710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaderna S, Kropff B, Kodel Y, Shen S, Coley S, Lu S, Britt W, Mach M. Deletion of gpUL132, a structural component of human cytomegalovirus, results in impaired virus replication in fibroblasts. J Virol. 2005;79:11837–11847. doi: 10.1128/JVI.79.18.11837-11847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton KR, Durch JS, Lawrence RS. Vaccines for the 21st century: a tool for decision making. Baltimore, MD: National Academy of Sciences Press; 1999. [PubMed] [Google Scholar]
- Vogel P, Weigler BJ, Kerr H, Hendrickx AG, Barry PA. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab Anim Sci. 1994;44:25–30. [PubMed] [Google Scholar]
- Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A. 2005;102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills MR, Ashiru O, Reeves MB, Okecha G, Trowsdale J, Tomasec P, Wilkinson GW, Sinclair J, Sissons JG. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J Immunol. 2005;175:7457–7465. doi: 10.4049/jimmunol.175.11.7457. [DOI] [PubMed] [Google Scholar]
- Wussow F, Yue Y, Martinez J, Deere JD, Longmate J, Herrmann A, Barry PA, Diamond DJ. A vaccine based on the rhesus cytomegalovirus UL128 complex induces broadly neutralizing antibodies in rhesus macaques. J Virol. 2013;87:1322–1332. doi: 10.1128/JVI.01669-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Barry PA. Rhesus cytomegalovirus a nonhuman primate model for the study of human cytomegalovirus. Adv Virus Res. 2008;72:207–226. doi: 10.1016/S0065-3527(08)00405-3. [DOI] [PubMed] [Google Scholar]
- Yue Y, Kaur A, Eberhardt MK, Kassis N, Zhou SS, Tarantal AF, Barry PA. Immunogenicity and protective efficacy of DNA vaccines expressing rhesus cytomegalovirus glycoprotein B, phosphoprotein 65-2, and viral interleukin-10 in rhesus macaques. J Virol. 2007;81:1095–1109. doi: 10.1128/JVI.01708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.