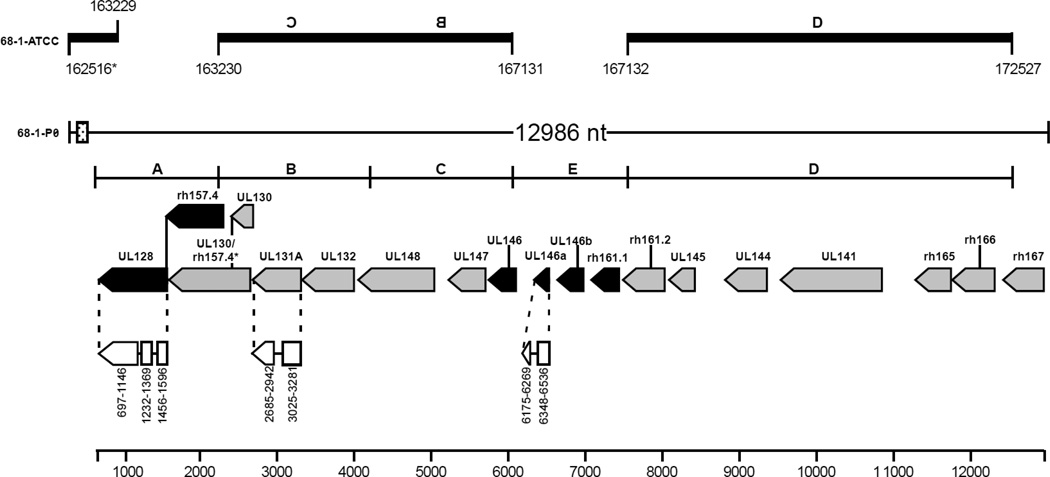
Fig. 1.
Genomic arrangement of UL/b’ open reading frames (ORFs) sequenced from the original source of RhCMV 68-1-P0 compared with passaged RhCMV 68-1 deposited in the ATCC (68-1-ATCC). Nucleotide numbering of RhCMV 68-1-ATCC (based on Rivailler et al., 2006) is shown at the top for comparison. The genomic structures, consensus regions (A-D), and predicted ORFs are depicted based on previously reported sequences of RhCMVs from low passage or unpassaged isolates (Rivailler et al., 2006; Oxford et al., 2008). RhCMV 68-1-P0 ORFs are on the complementary strand. Gray shaded ORFs are present or have a highly homologous region in 68-1-ATCC, while black shaded ORFs are absent from 68-1-ATCC. White regions joined by lines are proposed of exons. The splice variant for UL130 is indicated as UL130/rh157.4. A putative microRNA region (22-mer repeated 18 times, dotted box) is highlighted at the 5’ end of the contig and is also present in the 68-1-ATCC strain. The figure is based on that from Oxford et al. 2008.