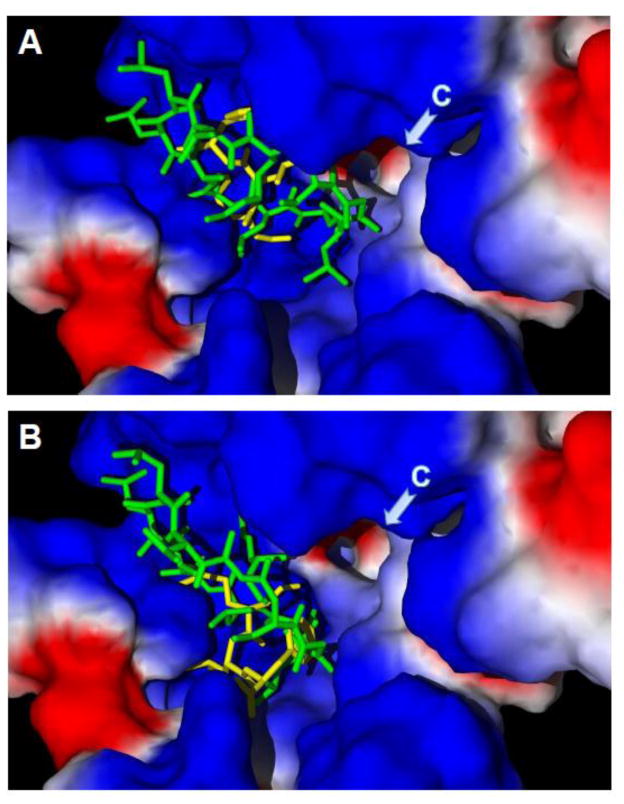
Figure 2.
Docking poses of cyclosporine H and fMLF into the FPR1 ligand binding site. Docking poses for CsH (green) and fMLF (yellow) were determined using Molegro software for the Val101 (Panel A) and Leu101 (Panel B) FPR1 variants. Briefly, the PDB structure of a rhodopsin-based homology model of FPR1 was imported into the Molegro program. To perform “mutation,” Leu101 was changed into Val101, and the latter was optimized using a built-in module of the program. The search space was defined as a sphere of 11 Å, as described previously [210]. Side chains of 34 residues inside this sphere were treated as flexible during docking. CsH and fMLF structures were prepared using the HyperChem 7 package and optimized with the MM+ force field. Arrows indicate channel C (region designated as previously reported [210]). Surface coloring is based on electrostatic properties, where negatively and positively charged areas are shown in red and blue, respectively.