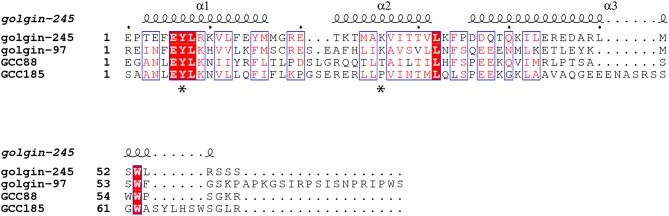
Figure 2.
Sequence alignment of the GRIP domains of human Golgin-245, Golgin-97, GCC88, and GCC185 based on the crystal structure of Golgin-245 generated from PROMALS3D (Pei et al., 2008) and ESPript 3.0 (Robert and Gouet, 2014). The secondary structures of human Golgin-245 are shown above. Invariant residues are in white with red background; similar residues (with global scores between 0.7 and 1.0) are in red, framed in blue, and other residues are in black. Asterisks indicate the invariant tyrosine that is critical for Golgin-245 and Golgin-97 localization and Arl1 binding, as well as the proline in GCC185 that may break helix 2.