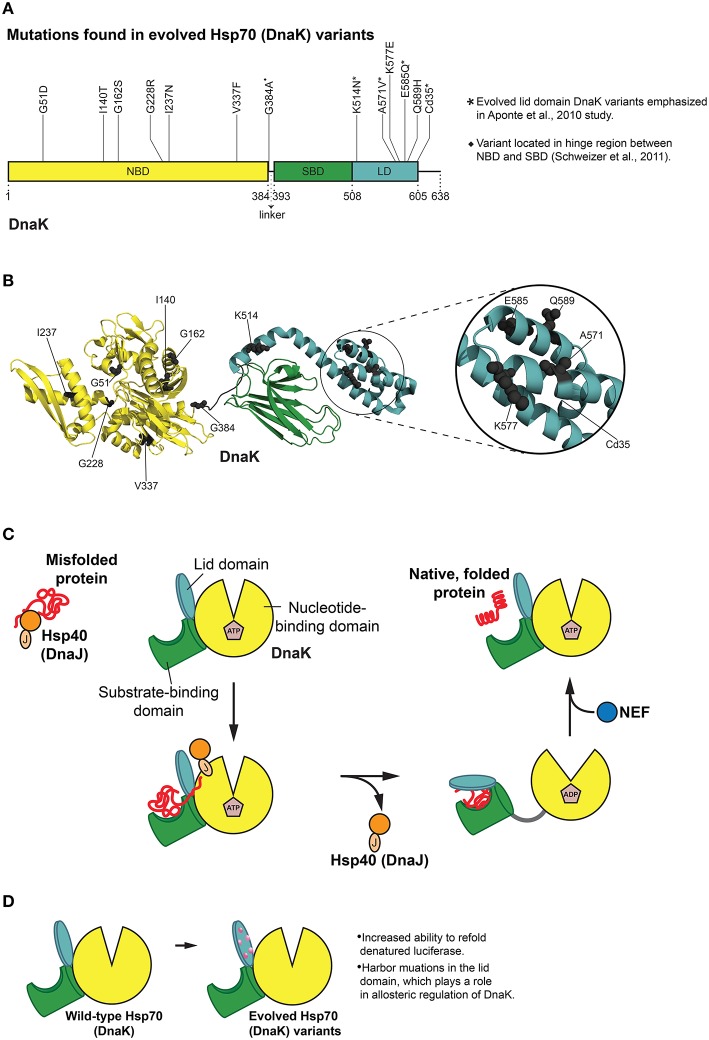
Figure 4.
Hsp70 (DnaK) protein folding cycle and features of evolved variants. (A) Mutations in evolved DnaK variants localize to the nucleotide binding domain (NBD) and lid domain (LD) of DnaK, rather than the substrate binding domain (SBD; Aponte et al., 2010). Interestingly, one enhanced variant harbors a C-terminal truncation of 35 amino acids (Cd35; Aponte et al., 2010). (*) Denotes evolved lid domain DnaK variants emphasized in Aponte et al. (2010) study. (♦) Denotes G384A mutation isolated in subsequent evolution of Cd35 DnaK variant (Schweizer et al., 2011). Based on Figure 2 in Aponte et al. (2010). (B) Structure (PDB ID: 2KHO) of ADP-bound DnaK with a 33 residue C-terminal truncation (Bertelsen et al., 2009) showing the positions of mutations (gray spheres) in evolved DnaK variants. Colors correspond to (A). Based on Figure 2 in Aponte et al. (2010). (C) DnaK protein folding cycle. Hsp40 (DnaJ) binds and presents substrate to DnaK. DnaK then weakly interacts with substrate in proximity to DnaJ, and DnaJ stimulates ATPase activity of DnaK thus stabilizing substrate binding. A nucleotide exchange factor (NEF) promotes nucleotide exchange, Hsp70 returns to the ATP-bound state and substrate is released (Goloubinoff and De Los Rios, 2007; Kim et al., 2013). (D) Features of evolved DnaK variants. Evolved DnaK variants show increased ability to refold denatured luciferase, and contain mutations in the lid domain, which plays a role in allosteric regulation of DnaK (Jiang et al., 2005; Vogel et al., 2006; Swain et al., 2007; Aponte et al., 2010).