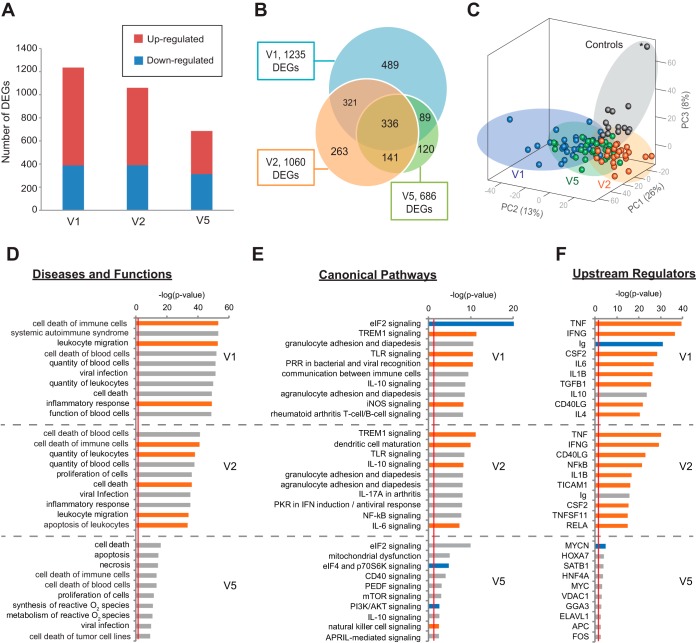
FIG 2 .
Longitudinal differential gene expression and pathway analysis of Lyme disease. (A) Bar chart of the numbers of genes found to be upregulated or downregulated at Lyme disease diagnosis (V1), 3 weeks post-treatment (V2, after a standard course of antibiotics), and 6 months post-treatment (V5). (B) Venn diagram representing the number of DEGs between Lyme disease patients and controls at three time points. (C) Principal component analysis (PCA) of Lyme disease patients and controls at three time points on the basis of 1,759 unique DEGs identified at V1, V2, and V5. The asterisk represents a subject in the control group who looks like an outlier in the PCA plot but is not shown to be an outlier by PCA analysis of the control samples (see Fig. S2 in the supplemental material). Note that the PC3 axis in the PCA plot accounts for only 8% of the variance in the data set. (D to F) Top 10 disease and functional categories (D), top 10 canonical pathways (E), and top 10 upstream regulators (excluding drug categories) (F) predicted to be involved in Lyme disease at (V1, V2, and V5) with categories, pathways, and genes ranked by the negative log of the P value of the enrichment score. The color scheme is based on Z scores, with activation in orange, inhibition in blue, and undetermined directionality in gray. The red line represents the designated significance threshold (P < 0.05).