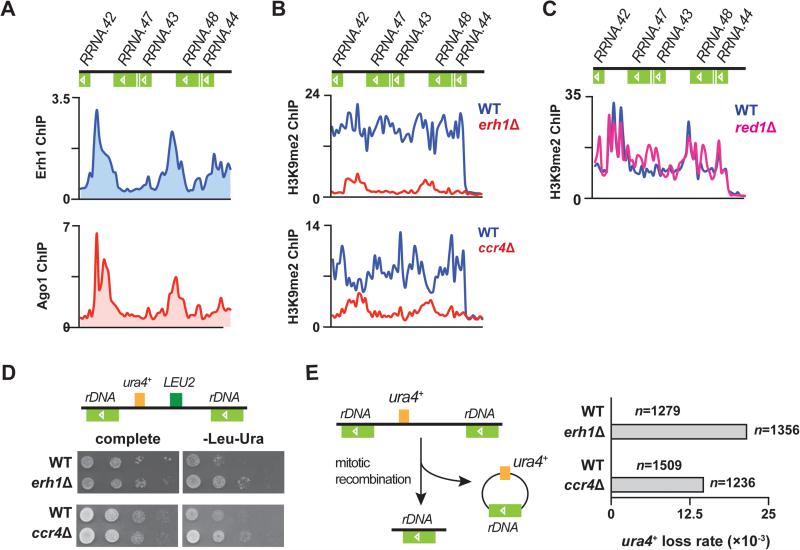
Figure 6. Erh1 and Ccr4 regulate heterochromatin formation at rDNA repeats to protect rDNA integrity.
(A) ChIP-chip of Erh1-GFP (top) and myc-Ago1 (bottom) at rDNA repeats in WT. (B) ChIP-chip of H3K9me2 at rDNA repeats in WT and erh1Δ (top), and in WT and ccr4Δ (bottom). (C) ChIP-chip of H3K9me2 at rDNA repeats in WT and red1Δ. (D) LEU2 and ura4+ markers inserted at rDNA repeats (Ylp2.4pUCura4+-7) are de-repressed in erh1Δ and ccr4Δ. A schematic representation of Ylp2.4pUCura4+-7 is shown (top). WT and erh1Δ (middle) or WT and ccr4Δ (bottom) carrying Ylp2.4pUCura4+-7 were spotted onto complete medium or minimal medium lacking leucine and uracil (-Leu-Ura) and incubated at 30°C for 2 days. (E) Increased frequency of mitotic recombination at tandem rDNA repeats in erh1Δ and ccr4Δ. A schematic representation of rDNA::ura4+ loss is shown (left). WT, erh1Δ and ccr4Δ carrying Ylp2.4pUCura4+-7 were first plated on medium lacking leucine, transferred to complete medium, and then replica plated to medium lacking uracil (-Ura). Mitotic recombination at rDNA loci was indicated by the lack of growth on -Ura plates, and was confirmed by genomic PCR. “n” indicates the number of cells counted for each strain. See also Figure S6.