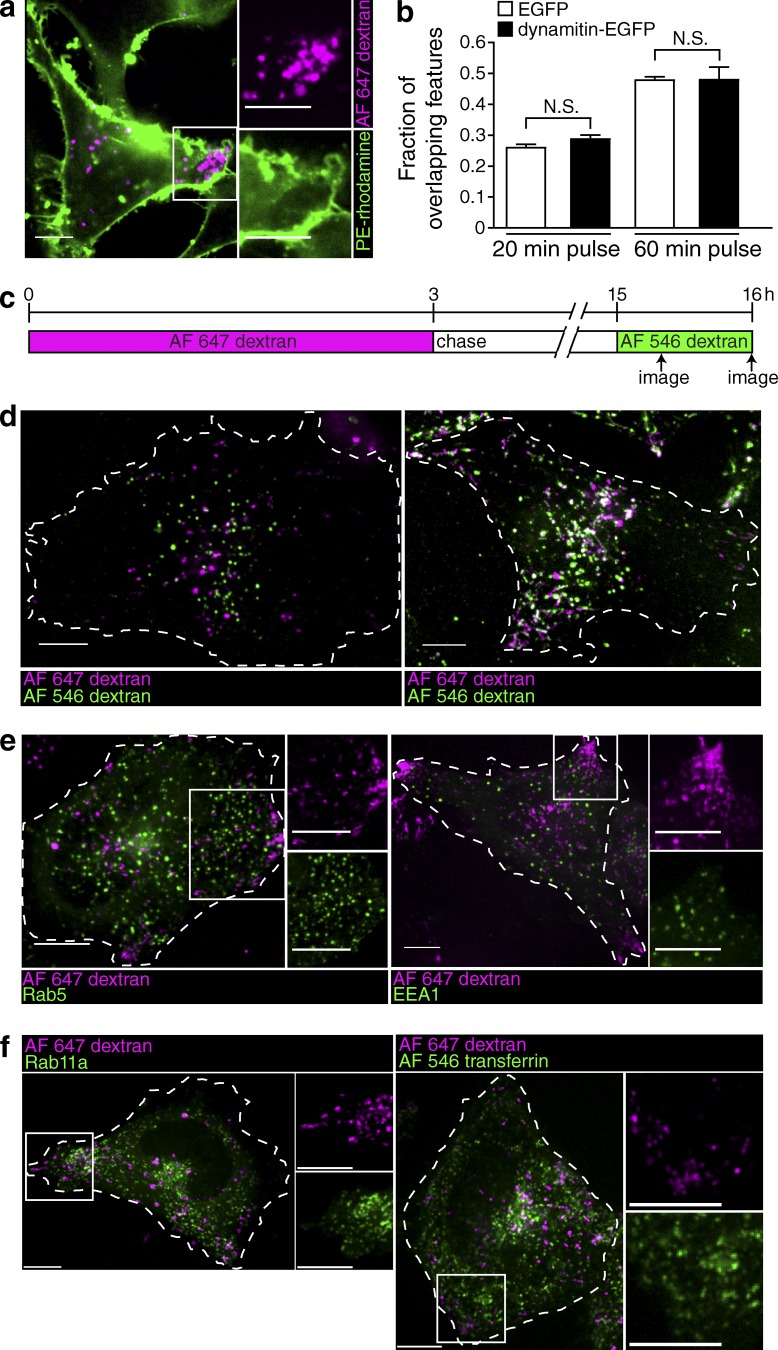
Figure 5.
Peripheral lysosomes do not exhibit enhanced interaction with the plasma membrane, nor do they colocalize with early or recycling endocytic markers. (a) The lysosomes of HeLa cells were loaded with Alexa Fluor 647–dextran (magenta) and transfected with dynamitin-EGFP cDNA (not depicted). The plasma membrane was stained with the fluorescent marker PE-rhodamine (green), and the cells were imaged by confocal microscopy in the 5–10-min interval after membrane labeling. The white box indicates the region magnified in the right-hand insets. (b) The fraction of overlapping features (Alexa Fluor 647–dextran-positive lysosomes containing Alexa Fluor 546–dextran) at 20 and 60 min was determined in lysosomes of both control (EGFP transfected) and dynamitin-EGFP–transfected cells. Error bars represent SEM (n = 6 cells for each condition from three independent experiments). (c) Diagrammatic presentation of the protocol used: lysosomes were loaded with Alexa Fluor 647–dextran for 3 h, chased for 12 h, and then loaded with Alexa Fluor 546–dextran for 20 or 60 min. Cells were imaged by confocal microscopy when indicated by the arrows. (d) Representative images of control cells imaged 20 min (left) or 60 min (right) after addition of Alexa Fluor 546–dextran. Cells of interest are outlined with dashed lines. (e) Lysosomes were loaded with Alexa Fluor 647–dextran (magenta) and transfected with Rab5-EGFP (green) or EEA1-EGFP (green). Cells were imaged by confocal microscopy. White boxes indicate the regions magnified in the right-hand insets. (f) Lysosomes were loaded with Alexa Fluor 647–dextran (magenta) and transfected with Rab11a (green) or loaded for 20 min with Alexa Fluor 546–transferrin (green). Cells were imaged by confocal microscopy. White boxes indicate the regions magnified in the right-hand insets. Bars, 10 µm.