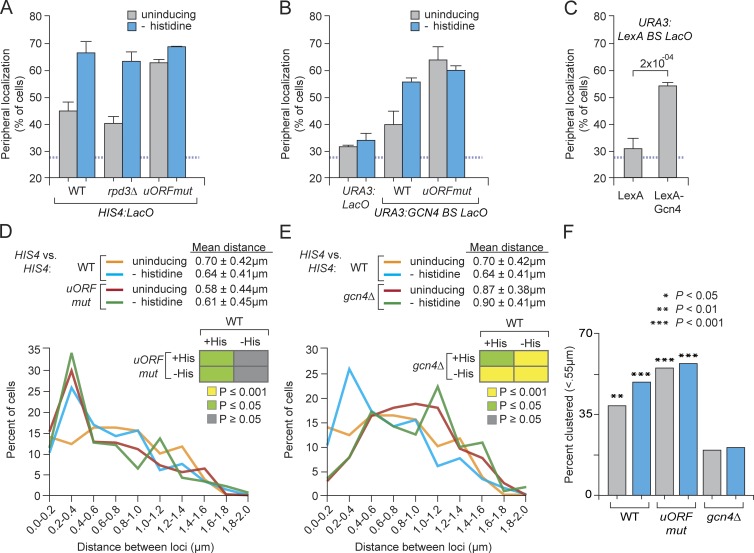
Figure 8.
Translational control of Gcn4-mediated peripheral targeting and interchromosomal clustering. (A–C) Peripheral localization of HIS4 (A), URA3 (B), URA3:GCN4 BS (B), and URA3:LexA BS (C) in the indicated strains grown under uninducing (gray bars) or inducing (cyan bars) conditions. Mutations in the uORFs were introduced into the chromosomal GCN4 locus (Methods). *, P ≤ 0.05 (Fisher exact test) between uninducing and inducing condition. WT, wild-type. (C) Peripheral localization of URA3-LexA BS in strains expressing LexA or LexA-Gcn4. Mean and SEM from three of more biological replicates (30–50 cells per replicate; p-value from Fisher exact test). (D and E) Distribution of distances, binned into 0.2-µm bins, between alleles of HIS4 in wild type, GCN4uORF (D), or gcn4Δ (E) homozygous diploid strains grown ± histidine. (insets) P-values from Wilcoxon rank sum test for each comparison. (F) Fraction of cells in which the two alleles of HIS4 were ≤0.55 µm apart (p-values from Fisher exact test compared with the gcn4Δ strain under uninducing conditions).