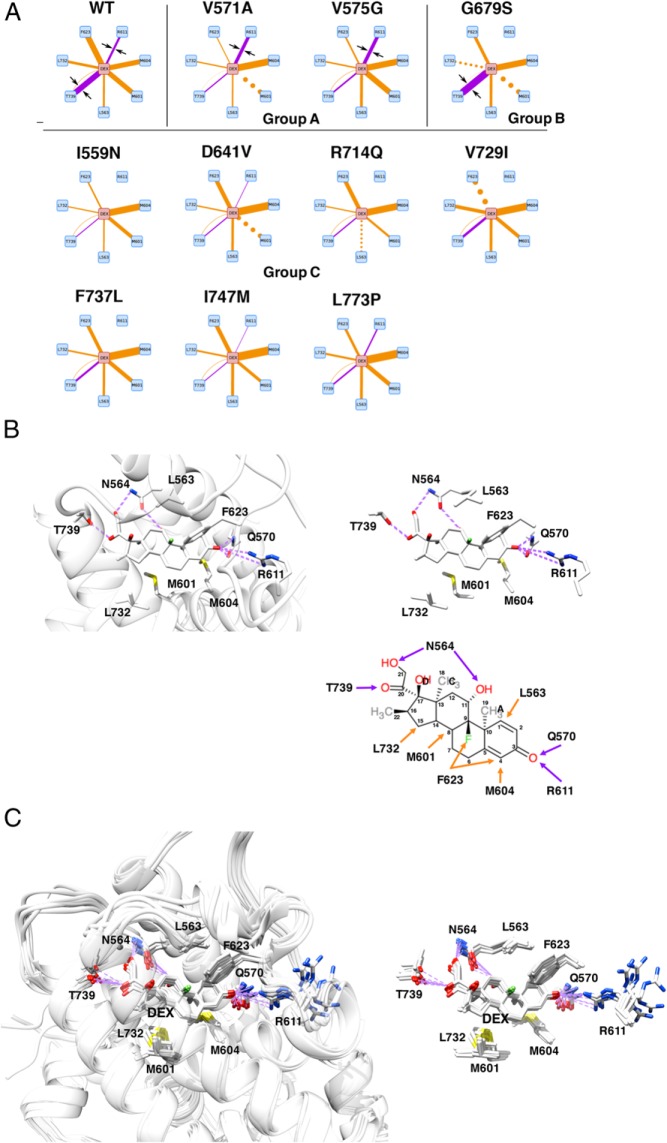
Figure 5.
Alteration of the electrostatic bond formed by R611 and T739 of pathologic GRα mutants may largely explain the reduced affinity of many pathologic GRα mutants to this steroid. A, Diagram of individual molecular interactions between dexamethasone and wild-type (WT) GRα or pathologic GRα mutants. Purple and orange lines indicate electrostatic and noncovalent bonds, respectively. Line thickness indicates frequency of the observed interaction (thus strength of interaction), whereas dotted lines indicate no statistical change in the frequency of interaction compared with WT GRα. Only the interactions dramatically altered in the mutant receptors are shown. Arrows are for attracting readers' attention. Based on the strength of interaction created by R611 and T739 of receptors, pathologic GRα mutants can be categorized into 3 groups (groups A, B, and C). Full interaction diagrams are shown in Supplemental Figure 4. B, 3D or schematic models (residues only) of the molecular interaction between WT GRα LBP and dexamethasone. Top left and top right panels demonstrate 3D interaction images of dexamethasone and the key residues of WT GRα that create important contacts to this steroid. Bottom right panel shows schematic molecular interaction between WT GRα and dexamethasone. Purple and orange arrows indicate electrostatic and noncovalent bonds, respectively. C, Superimposed 3D images of dexamethasone and the key residues of all pathologic GRα mutants. Panels demonstrate superimposed 3D interaction images of dexamethasone and the key residues of all pathologic GRα mutants. Among the key amino acids of pathologic mutants participating in interaction with dexamethasone, R611 is largely deviated in these mutant receptors, which underlies reduced/disappeared electrostatic interaction between this residue and carbonyl oxygen at carbon-3 of dexamethasone. Q570 and N564 are omitted from these panels. DEX, dexamethasone; WT, wild type.