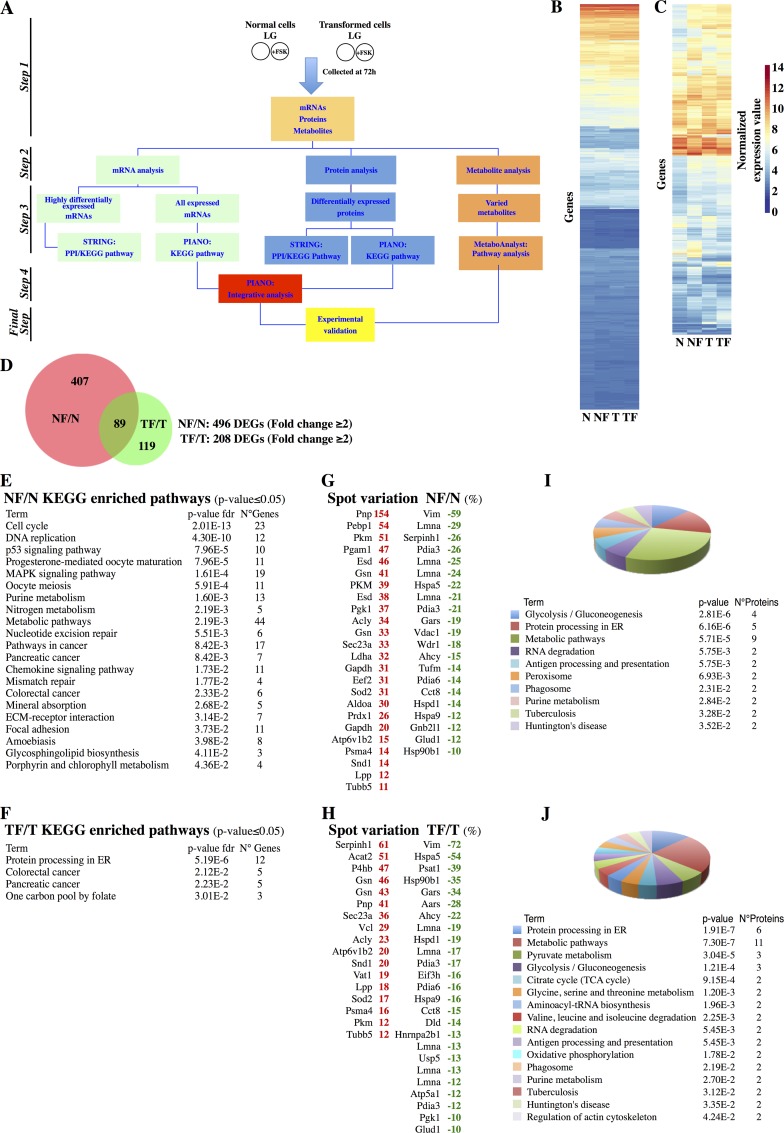
Fig 1. Transcriptomic and proteomic analysis of Normal and Transformed cells treated with FSK.
(A) Workflow followed to analyze the transcriptomic, proteomic and metabolomic data. (B-C) Hierarchical clustering of 17669 differentially expressed genes of Normal cells and Transformed cells -/+ FSK (B) and 615 genes having fold change ≥2 in the two comparisons NF/N and TF/T (C). (D) Venn diagram representing the distribution of the 615 genes in the two comparisons. (E-F) Enriched KEGG pathways obtained by STRING analysis of the 496 and 208 DEGs in the comparisons NF/N (E) and TF/T (F) (fold change ≥2). (G-H) In red the up-regulated proteins and in green the down-regulated ones identified in proteomic analysis in the comparison NF/N (G) and TF/T (H). (I-J) Enriched KEGG pathways obtained by STRING analysis of the differentially expressed proteins in the two comparisons NF/N (I) and TF/T (J). The dimension of the pie slice is referred to the number of proteins associated to each pathway. The bottom list represents the identified pathways, depicted by using a color code, the number of proteins belonging to the pathway and the p-value (p-value <0.05).