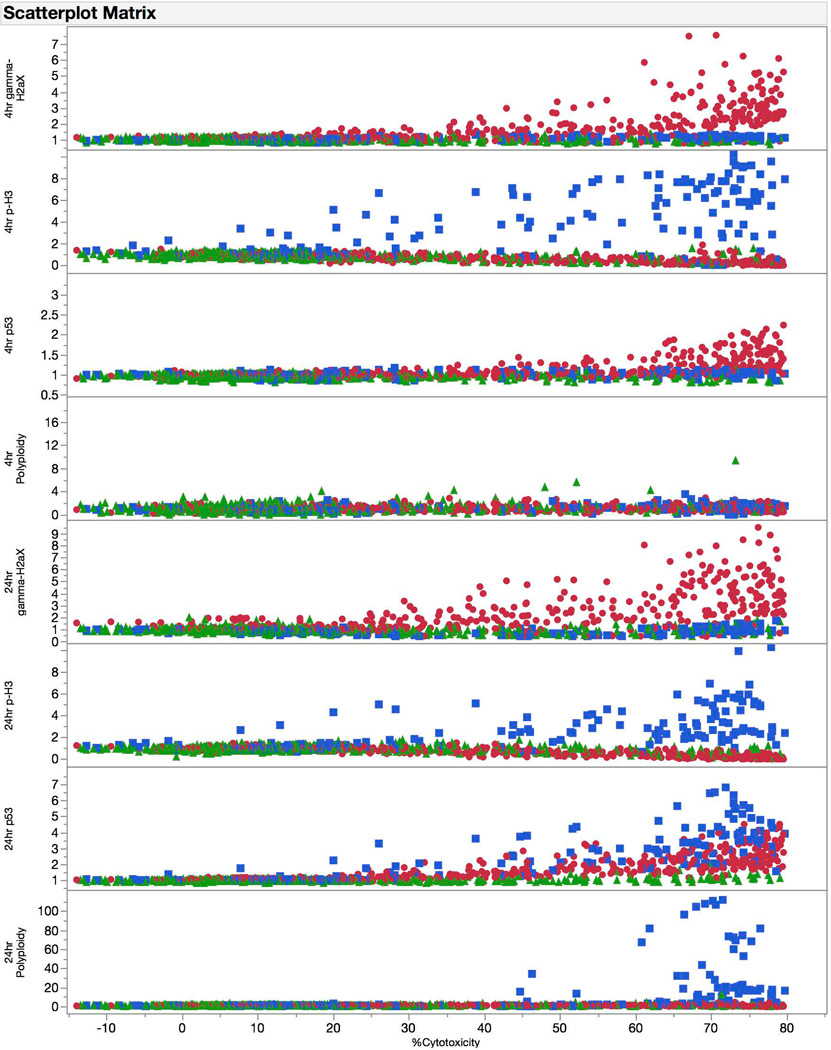
Figure 3.
Fold-increase responses for each biomarker at each of two time points are graphed against a measure of cytotoxicity (i.e., 100% - Relative Nuclei Count at 24 hrs). The graphs are aggregate data for 67 training set chemicals, and are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. These graphs provide insight into which biomarkers are effective at discriminating among chemical classes, and provide further evidence that testing up to cytotoxic concentrations, to a point, maximizes signal to noise ratios across these disparate endpoints.