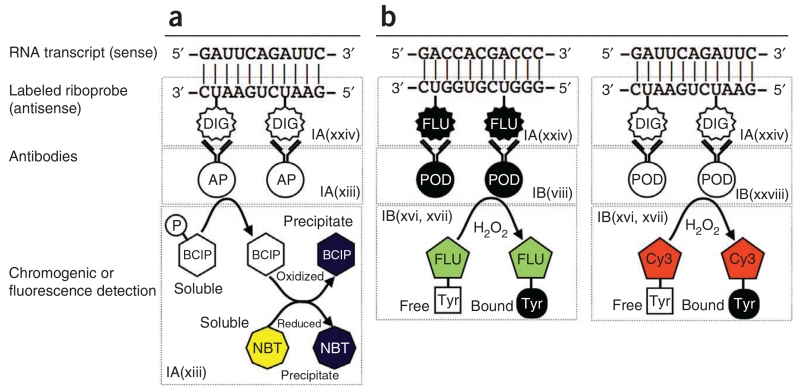
Figure 1.
Principal steps in chromogenic ISH and FISH. (a,b) Chromogenic ISH is shown in a, FISH in b. As described in the text, fluorescence is preferable for simultaneous detection of multiple RNA transcripts in the same cell. Both approaches use an antisense riboprobe in which some of the uridine residues are labeled with either digoxigenin (DIG) or fluorescein (FLU). Once the probe is bound to the target transcript, the digoxigenin or fluorescein molecules are then recognized by the appropriate antibodies. The antibodies are conjugated to either alkaline phosphatase (AP; in the case of chromogenic detection) or peroxidase (POD; in the case of fluorescence detection). In chromogenic detection, the presence of alkaline phosphatase is then visualized by BCIP and NBT. The alkaline phosphatase removes a phosphate group from BCIP, which is a colorless soluble compound. The BCIP then becomes oxidized in a reaction with NBT (a soluble yellow compound), which is simultaneously reduced. Both molecules precipitate out of the solution and assume a dark purple color. In fluorescence detection, numerous tyramide-conjugated fluorophores (e.g., Cy3 or fluorescein) aggregate in the vicinity of the bound RNA probe, as the tyramide is activated by the peroxidase (POD) bound to the antibody.