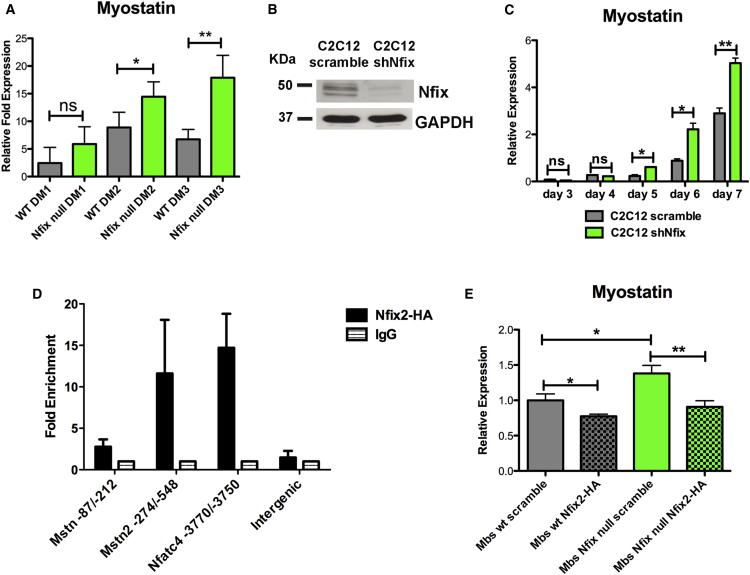
Figure 5.
Nfix Regulates Myostatin Expression in Differentiating Myoblasts through a Direct Binding to its Promoter
(A) Real-time qPCR showing Myostatin upregulation in differentiating SC-derived myoblasts. The SCs were isolated by FACS and plated in differentiation medium for a time course analysis from 1 to 3 days (DM1, DM2, and DM3) (n = 4 DM1 WT, n = 2 DM1 Nfix-null, n = 5 DM2 WT, n = 3 DM2 Nfix-null, n = 4 DM3 WT, and n = 4 DM3 Nfix-null). The data are presented as mean ± SD (not significant, ns; ∗p < 0.05; ∗∗p < 0.01; and two-tailed unpaired t test).
(B) Western blot analysis of Nfix expression in C2C12 myoblasts transduced with a control vector (C2C12 scramble) or with a vector carrying a shRNA targeting Nfix (C2C12 shNfix). GAPDH was used to normalize.
(C) Real-time qPCR analysis of Myostatin expression in scramble and shNfix C2C12 in a time course from 3 to 7 days in differentiation medium. The values are plotted as relative expression and normalized to GAPDH (n = 3 independent samples for each time point). The data are presented as mean ± SD (not significant, ns; ∗p < 0.05; ∗∗p < 0.01; and two-tailed unpaired t test).
(D) ChIP on differentiated C2C12 transduced with a vector expressing a HA-tagged Nfix2 isoform to test binding to putative Nfix binding sites on Myostatin promoter located at −87/−212 bp and −274/−548 bp from transcription start site. Binding on Nfatc4 promoter and on an intergenic region were used as positive and negative controls, respectively. The data are means of two independent experiments and expressed as fold enrichment (mean ± SD) relative to the IgG signal (n = 3 independent ChIP).
(E) Real-time qPCR for Myostatin in WT and Nfix-null SC-derived myoblasts transduced with a control vector (scramble) or with a vector overexpressing the Nfix2 isoform (Nfix2-HA). The values are plotted as relative expression and normalized to GAPDH (n = 3 samples for each group). The data are presented as mean ± SD (∗p < 0.05; ∗∗p < 0.01; and two-tailed unpaired t test).