Figure 1. BabAAD Interacts with Lewis b bg Antigens.
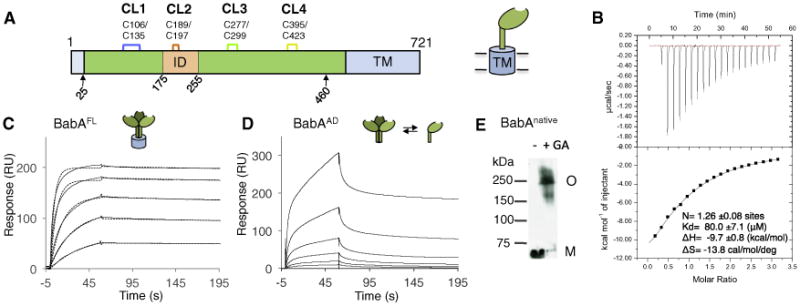
(A) Schematic of the BabA architecture. Arrows indicate the aa 25–460 BabA adhesin domain fragment (BabAAD). Abbreviations: CL, cysteine-clasped loops; TM, predicted transmembrane domain; ID, Bab insertion domain (Figures S2 and S3).
(B) ITC injection heats (upper) and normalized binding isotherm (lower) of the BabAAD titrated with Leb5.
(C) SPR sensorgram of full-length BabA (solid and dashed lines show raw and fitted binding curves, respectively, for 500, 250, 125, 62.5, 31.3, and 15.7 nM BabA, from the top down; with a dissociation constant Kd = 3.9E-10 ± 0.9E-10 (M), an association rate constant ka = 6.1E5 ± 1.4E5 (M−1 s−1), and slow dissociation rate constant, kd = 2.3E-4 ± 0.8E-4 (s−1).
(D) Similar SPR sensorgram of purified BabAAD binding to a Leb-coated chip; [BabA] as in (C).
(E) Immunoblot detection of BabA from glutaraldehyde(GA) crosslinked H.pylori 17875/Leb bacterial cells; M, monomer, O, BabA oligomer. See also Figure S1.
