FIGURE 1.
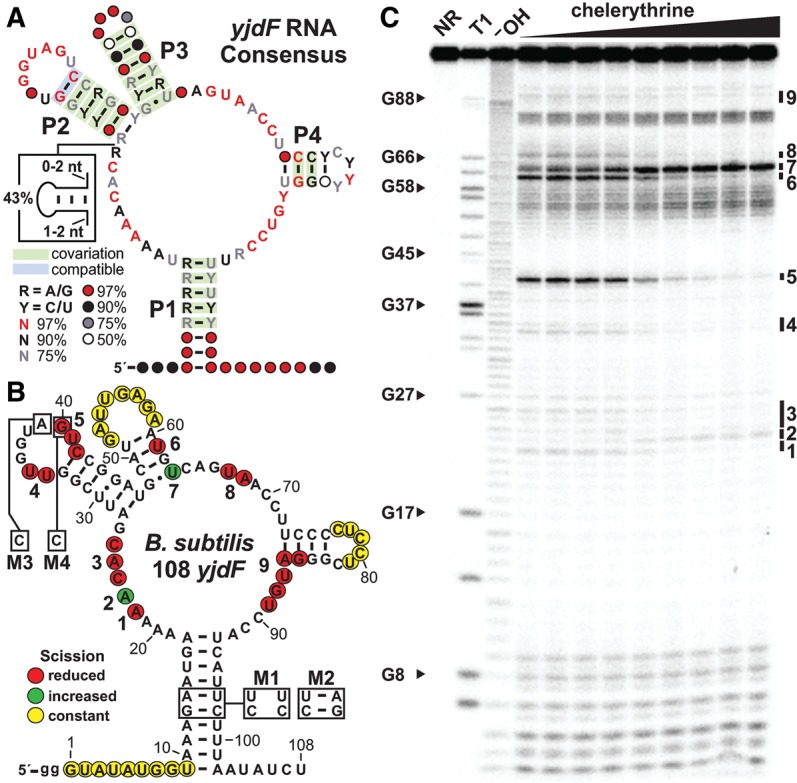
yjdF motif RNAs exhibit biochemical characteristics of riboswitch aptamers. (A) Consensus sequence and structure of yjdF motif RNAs based on 1060 distinct RNA representatives. Base-paired substructures P1 through P4, and sometimes an additional variable-sequence stem (box), are interspersed among regions of high sequence conservation. The percentage of representatives carrying the nucleotides depicted are indicated by colored letters (R is G or A, Y is C or U) or circles (designating the presence of any nucleotide), and colored boxes indicate there is evidence for natural sequence covariation or compatible mutation that retains base-pairing. (B) Sequence and secondary structure of the 108 yjdF RNA construct based on the 5′ UTR of the yjdF gene from Bacillus subtilis. The nucleotides altered in mutated RNA constructs M1, M2, M3, and M4 are depicted in boxes. Regions that undergo RNA strand scission as revealed by the in-line probing data depicted in C are identified with colored circles based on their characteristics. Nine regions that undergo increased or decreased scission are numbers 1–9. (C) Polyacrylamide gel electrophoresis (PAGE) analysis of in-line probing reactions of the 5′ 32P-labeled 108 yjdF construct with concentrations of the natural alkaloid drug compound chelerythrine ranging from 0 to 300 nM. NR, T1, and −OH designate no reaction, partial digestion with RNase T1, and partial digestion with alkali, respectively. Select bands corresponding to RNase T1 digestion after G residues are labeled according to the numbering system in B. Numbers 1–9 designate changes in banding patterns at the sites denoted in B.
