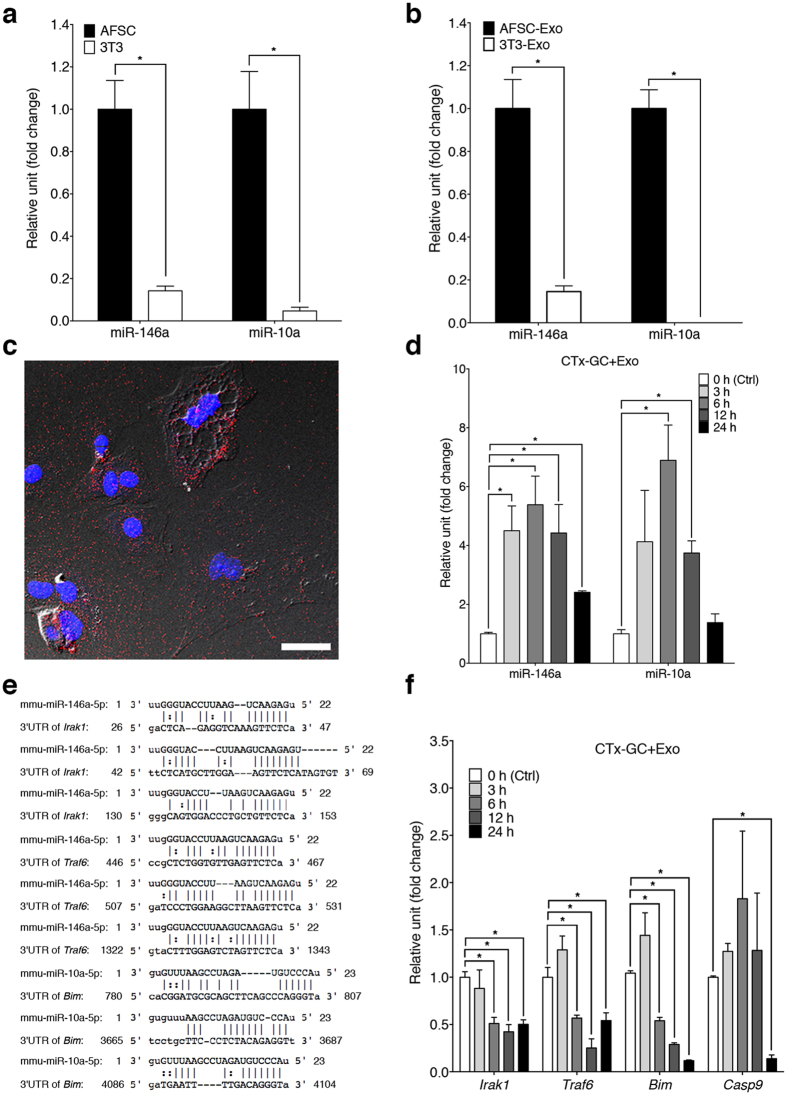
Figure 1. Delivery of miRNAs into damaged GCs via exosomes.
(a,b) qRT-PCR analysis of expression levels of miR-146a and miR-10a in AFSCs and NIH-3T3 (a) and their corresponding exosomes (b). Error bars represent s.e.m. n = 3. *P < 0.05; unpaired t test. (c) A representative micrograph shows that PKH26 labeled-AFSC-derived exosomes (30 μg ml−1 of exosomes proteins) (red) were incorporated into the cytoplasm of damaged GCs. Nucleus was stained by DAPI (blue). Scale bar, 40 μm. (d) The levels of miR-146a and miR-10a in damaged GCs cultured with AFSC-derived exosomes at different time points compared to 0 h (Ctrl). Error bars represent s.e.m. n = 3. *P < 0.05; unpaired t test. (e) The sequence alignment of miR-146a and its predicted target sites of the mouse Irak1 and Traf6 mRNA 3′-untranslated region (3′-UTR), and miR-10a and its putative target sites of the mouse Bim mRNA 3′-UTR. (f) The expression levels of Irak1, Traf6, Bim and Casp9 in damaged GCs cultured with AFSC-derived exosomes at different time points compared to 0 h (Ctrl). Error bars represent s.e.m. n = 3. *P < 0.05; unpaired t test.