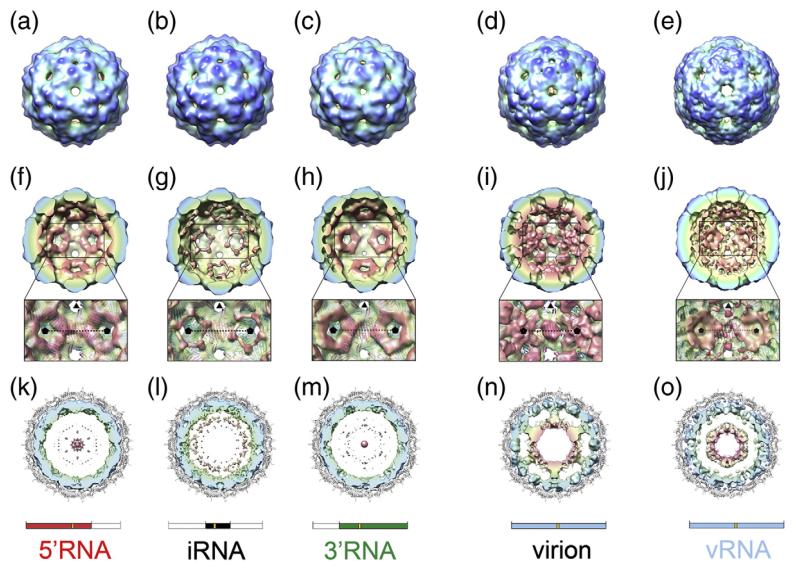
Fig. 5.
Cryo-EM structures of MS2 capsids. (a–e) Surface representation of the cryo-EM structures of MS2 capsids reassembled in the presence of 5′RNA (a), iRNA (b), 3′RNA (c) and vRNA (e). For comparison, the cryo-EM structure of the native virion is also shown (d).22 All structures have been Fourier filtered at ~15 Å resolution, and are radially coloured from blue at high radius to red at low radius. (f–j) The rear halves of the 3-D structures of 5′RNA (f), iRNA (g), 3′RNA (h), wt MS2 virion (i) and vRNA (j) with the same radial colouring scheme. The view for each structure is looking down an icosahedral 2-fold axis. The inset is an expanded portion of each structure showing details of the RNA density surrounding the 5-fold axes (indicated by black pentagons). Atomic coordinates (cartoon representation and coloured as in Fig. 1) are fit into a now semi-transparent density. There is little or no density for RNA beneath the C/C dimers (on the 2-fold axes). (k–o) Central, 40 Å thick cross-section views of 5′RNA (k), iRNA (l), 3′RNA (m), the wt MS2 virion (n) and vRNA (o). In each case, the view is along a 3-fold axis, and the density for CP has been masked away. The fitted atomic coordinates are shown as a grey cartoon. The unfiltered density for packaged RNA is coloured radially. The virion (n) shows an outer shell of RNA immediately beneath the protein capsid and a second shell of RNA at lower radii, connected to the outer shell along 5-fold axes. The inner-shell density is not seen in the sub-genomic RNA cryo-EM structures at an equivalent threshold level, and only very weak features are seen in the maps at lower radii (k–m). The inner shell reappears when the vRNA is packaged (o). Schematics for the RNA present in the structures shown in each column are shown for clarity.