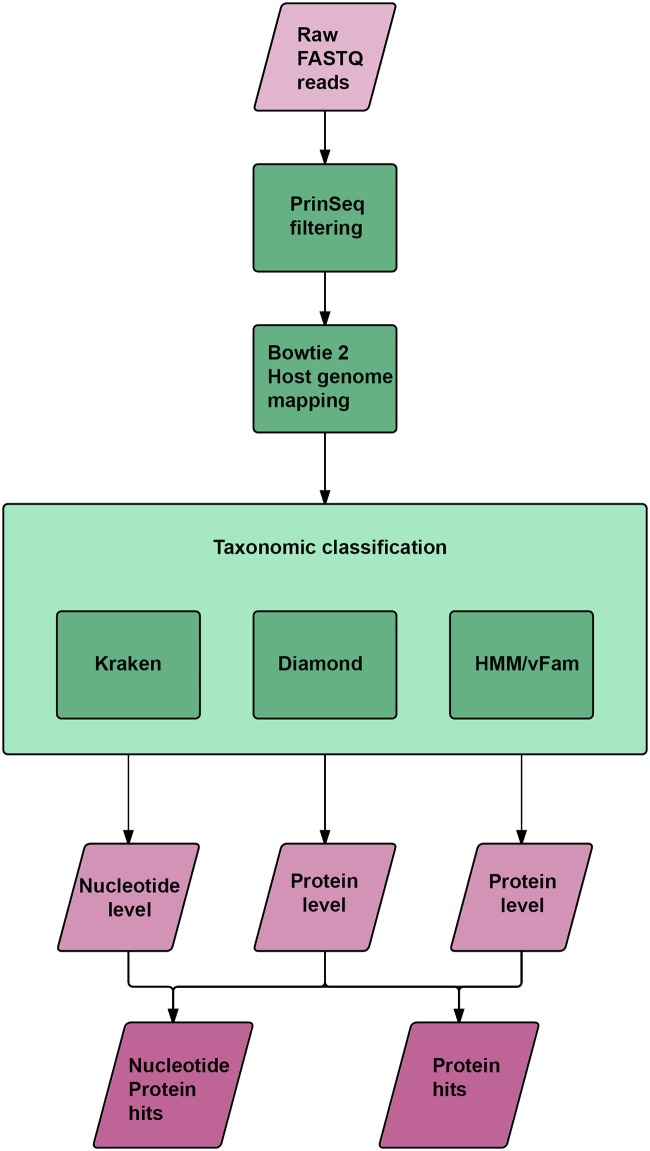
Fig 2. Schematic illustration of the bioinformatic pipeline.
Raw data from the sequencing platform, in FASTQ format, was processed by PrinSeq to select high quality reads. The fractions of high quality reads were then mapped towards the porcine genome, using Bowtie 2, to remove host sequences. The unmapped reads were then classified by three different methods, Kraken, Diamond and HMM/vFam. The resulting three datasets were grouped in two datasets, one representing reads classified on nucleotide and protein level and one representing reads classified only on protein level.