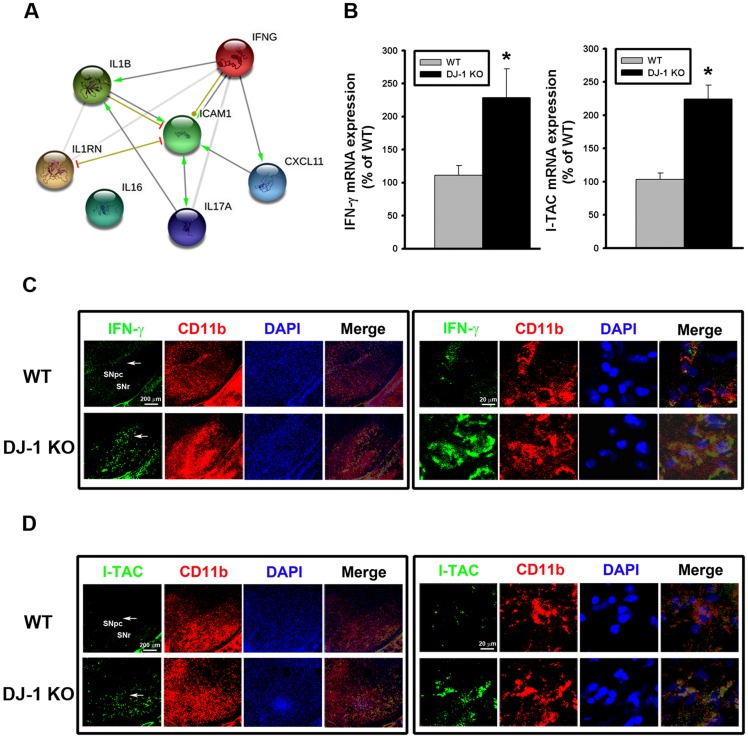
Fig 2. Up-regulation of IFN-γ and I-TAC in DJ-1 KO mice.
(A) IFN-γ-regulated biological network predicted from the bioinformatics tool STRING. IFNG (i.e. IFN-γ) was predicted to serve as a hub protein to control CXCL11 (i.e. I-TAC), IL-17, IL-1β, IL1RN, and ICAM1. (B) The basal mRNA levels of IFN-γ (left panel) and I-TAC (right panel) in substantia nigra were up-regulated in DJ-1 KO mice as compared with those in WT mice. (C) Increase of basal IFN-γ expression in microglia in substantia nigra of DJ-1 KO mice. The immunofluorescent labeling images of IFN-γ (green), CD11b (red) and DAPI (blue) in substantia nigra of WT (upper images) and DJ-1 KO (lower images) mice were shown. (D) Increase of basal I-TAC expression in microglia in substantia nigra of DJ-1 KO mice. The immunofluorescent labeling images of I-TAC (green), CD11b (red) and DAPI (blue) in substantia nigra of WT (upper images) and DJ-1 KO (lower images) mice were shown. The arrow indicates the brain area that is magnified and shown in the right panel. The merged image shows the co-localization of two proteins in microglia. Data were normalized as percentage of mean mRNA level in WT mice and presented as mean ± S.E.M. (n = 5 for each group) * p<0.05. SNpc: substantia nigra pars compacta; SNr: substantia nigra pars reticulate.