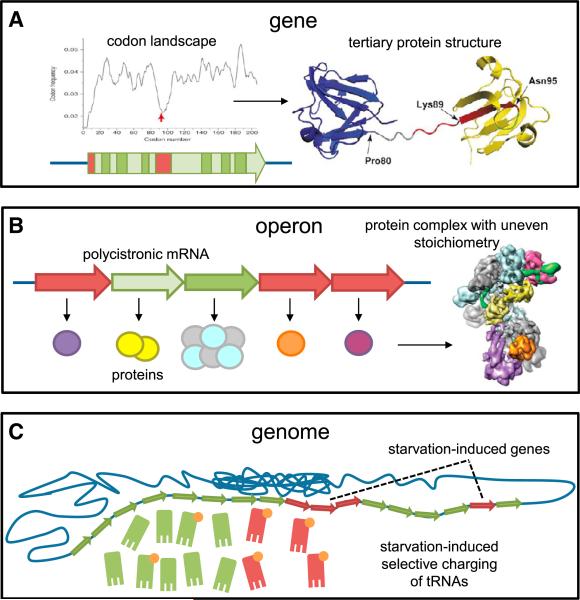
Figure 4. Codon Landscapes and Intergenic Codon Bias.
(A) The intragenic codon landscape can modulate the translation elongation rate, contributing to appropriate co-translational folding of the protein's secondary structure elements and domains. The red arrow indicates a stretch of rare codons encoding the red colored residues in the protein structure. Apart from a ramp sequence at 5′ end of the coding sequence, a valley in the codon landscape occurs between the two domains. The picture represents the bovine β-B2 crystallin and is adapted from Komar (2009).
(B) Differential expression of genes residing in an operon is controlled by differential translation. This is accomplished mainly by enhanced initiation (as reflected by a higher ribosome density) and, to some extent, by adjusted elongation (as reflected by a more optimal codon bias). An example is shown of the operon encoding the CRISPR-associated Cascade complex from E. coli with uneven stoichiometry; cryo-electron microscopic structure is from Wiedenheft et al. (2011).
(C) The expression of sets of functionally related genes can be co-regulated on the basis of their codon bias. The available pools of aminoacyl-tRNAs can specifically change under certain conditions, which lead to improved translation of genes with adapted codon bias. As an example, genes encoding amino acid biosynthesis pathways are shown, which contain many rare codons translated by tRNAs that remain highly charged during amino acid starvation.