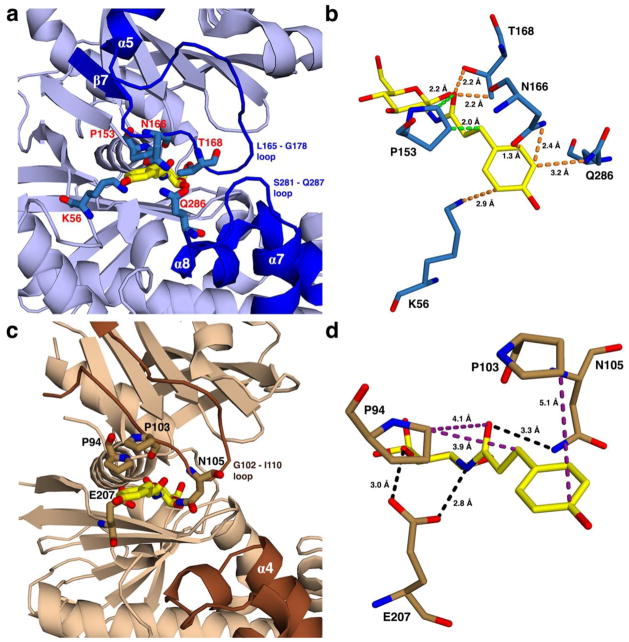
Figure 3.
Views showing the outer part of the active site for proposed HPOP-GlcN complexes of (a) HsHxKIV and (c) TcGlcK, from PDB entries 4MLE and 2Q2R, respectively. The HPOP-GlcN ligand was modeled into each kinase by superimposing its sugar moiety onto the glucose ligand, followed by removing the original glucose coordinates. Energy minimization was not applied in the modeling of HPOP-GlcN. Non-identical residues are indicated on the outer part of the active sites of both enzymes with residue labels as red for HsHxKIV (a) and black for TcGlcK (c). In these proposed complexes, panel (a) shows that various residue side chains of HsHxKIV (T168, N166, Q286, and K56) have the potential for steric clash with HPOP-GlcN while P153 has the highest likelihood of steric clash for the same ligand because the glucose moiety of HPOP-GlcN is in a fixed position. This situation is predicted to not allow HPOP-GlcN to bind in HsHxKIV. Panel (c) reveals minimum to no steric clashing effects present in the same region for TcGlcK. Panels (b) and (d) show HsHxKIV and TcGlcK on different viewing angles from panels (a) and (c), respectively. Protein residues are shown with very close contact interactions to HPOP-GlcN in HsHxKIV (b) and are essentially distant in TcGlcK (d). Interactions are pending that HPOP-GlcN is able to bind as the two glucose kinase/inhibitor models suggest and are color-coded as follows: orange represents close contacts from “flexible” residue side chains that are expected to move and avoid steric clashing effects; green represents close contacts from a rigid proline (P153) expected to have unavoidable steric clashing effects; purple represents relatively distant contact interactions; and black represents putative hydrogen bonding interactions.