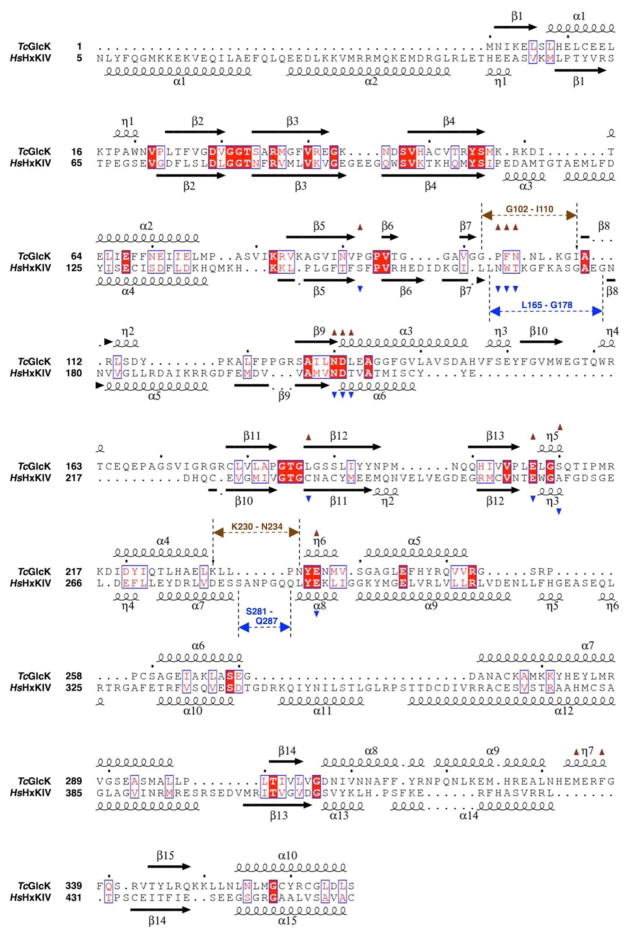
Figure 4.
Structure-based sequence alignment of TcGlcK (PDB entry 2Q2R) and HsHxKIV (PDB entry 3IDH) (34), as determined with the web-based protein structure comparison service Fold at European Bioinformatics Institute (PDBeFold) (35) and STRIDE (36, 37). The structural alignment was created with ESPript (version 3.0) (38). The overall sequence identity is 19% based on this alignment. The symbols α, η, and β indicate the secondary structural elements α-helices, 310-helices, and β-strands, respectively. Residues having exact identity are boxed and indicated by white letters on red background; residues with near identity are boxed and indicated by red letters on white background. Active site residues are indicated by brown triangles for TcGlcK and by blue triangles for HsHxKIV. The positions for two important loop segments are indicated by arrows having dashed lines (brown for TcGlcK and blue for HsHxKIV).