Figure 1. AKT genes signature co-regulated by EMT discriminates basal-like versus luminal subtype of breast cancers.
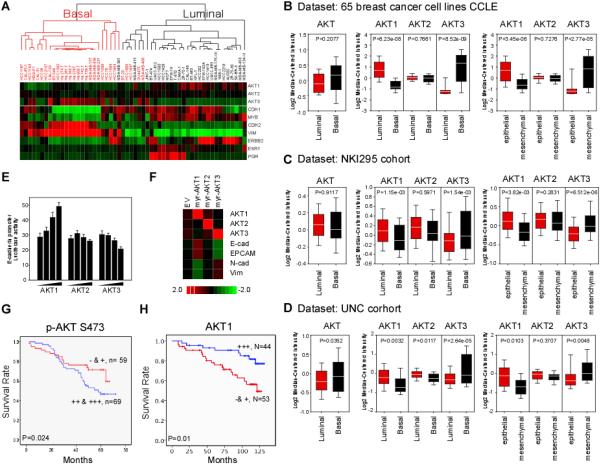
(A) Nonsupervised clustering of 54 breast cancer cell lines based on 10 genes. Gene expression heat map showing distinct expression pattern between basal- and luminal-breast cancer cell lines. TNBC gene signatures ESR1, ERBB2 and PgR are included.
(B) Box plots showing the average expression level of AKT1/2/3 or individual AKT isoforms gene in basal and luminal breast cancer cell lines (left) and (middle) from (A). Gene expression expressed as epithelial/mesenchymal ratio is shown (right).
(C and D) Box plots showing the average expression level of AKT1/2/3 (left), AKT1, AKT2 andAKT3 (middle and right) in basal and luminal cancers of NCI cohort (C) and of UNC cohort (D).
(E) Luciferase reporter assay of HEK-293 cells were transfected with the E-cadherin reporter together with increasing amount of myr-AKT1, myr-AKT2 or myr-AKT3.
(F) Gene expression heat map of E-cadherin, EPCAM, N-cadherin and vimentin in MDA-MB-231 cells expressing the myr-AKT isoforms.
(G) Kaplan-Meier analysis of p-AKT 473 survival of breast cancer patients in the MD Anderson Cancer Center cohort.
(H) Kaplan-Meier analysis of AKT1 by stratifying patients with quartiles partition.
