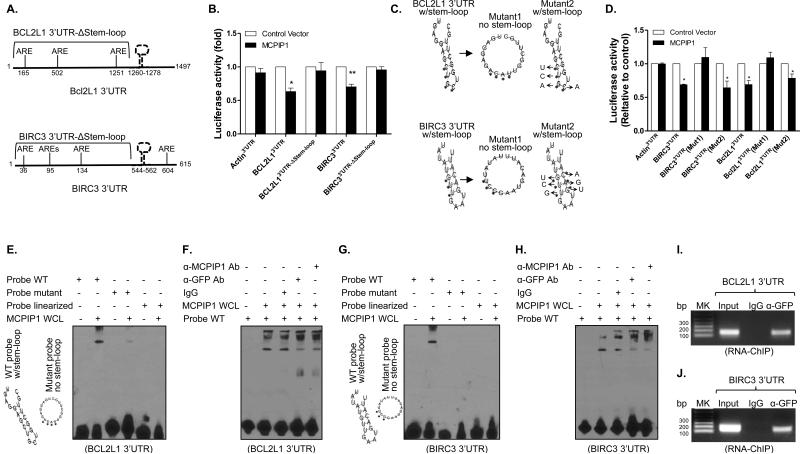
Figure 6.
MCPIP1 binds to stem-loop structure in the 3’UTR of anti-apoptotic genes for mRNA degradation. (A) Schematic representation of the luciferase reporter constructs of bcl2l1 and birc3 containing truncated 3’UTRs without the stem-loop (Δstem-loop). (B) HEK293 cells were co-transfected with luciferase reporters containing full-length 3’UTRs or truncated 3’UTRs (Δstem-loop) of bcl2l1 and birc3 in the presence of MCPIP1 or control vector. 48 hour later, luciferase activity was measured in cell lysates and shown as relative levels compared to cells transfected with empty vector. β-actin 3’UTR was used as negative control. Results shown represent the mean ± SD of four independent experiments. *: p< 0.05, **: p<0.01 between two groups. (C) The predicted stem-loop structures of Bcl2l1 (upper) and Birc3 (lower) in their 3’UTRs and mutation strategy (asterisks indicate base substitution). Mutant1 becomes unable to form stem-loop structure (middle) and Mutant2 still forms a stem-loop structure (left). (D) HEK293 cells were co-transfected with luciferase reporters containing full-length 3’UTRs or mutant 3’UTRs of bcl2l1 and birc3 in the presence of either MCPIP1 or control vector. Luciferase activity was measured in cell lysates and shown as relative levels compared to cells transfected with empty vector. Results shown represent the mean ± SD of four independent experiments (p< 0.05). (E) RNA-EMSA was performed with biotin-labeled probes corresponding to the BCL2L1 3’UTR, including WT probe, mutant probe and linearized probe, in the presence of whole cell lysates (WCL) extracted from MDA-MB-231/Tet-on cells after adding Dox. (F) WCL of MCPIP1-expressed MDA-MB-231/Tet-on cells were incubated with 1 ug of anti-MCPIP1 and anti-GFP antibody and IgG for 30 min, followed by performing RNA-EMSA assay as described above. (G & H) RNA-EMSA and supershift EMSA were respectively performed with biotin-labeled probes corresponding to the BIRC3 3’UTR, including WT probe, mutant probe and linearized probe, in the presence of whole cell lysates (WCL) extracted from MDA-MB-231/Tet-on cells after adding Dox. (I & J) RNA-ChIP was performed with genome fragments from MDA-MB-231/Tet-On cells 24 h after GFP/MCPIP1 induction. The pull-down 3’UTR sequences were amplified by PCR with primers flanking the putative stem-loop sequences.