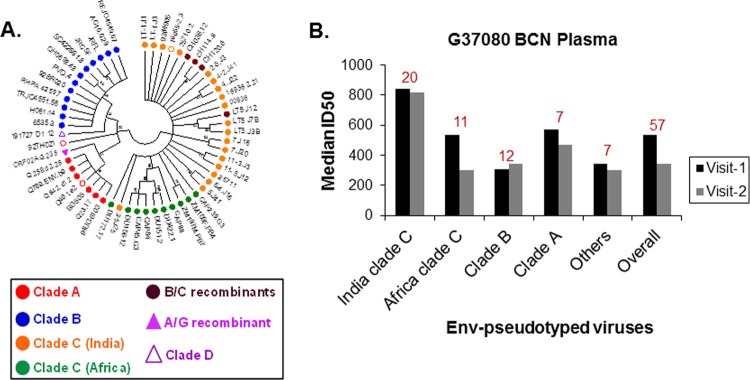
FIG 1.
(A) Genetic divergence of amino acid sequences of 57 HIV-1 Env (gp160) pseudoviruses used to assess neutralization breadth and potency of G37080 BCN plasma. The maximum likelihood bootstrapped consensus phylogenetic tree was constructed using the Jones-Taylor-Thornton (JTT) substitution model with 50 bootstrapped replicates in Mega 5.2. Bootstrapped values are shown at the nodes of each branch. Hollow circles represent envelopes (16055-2.3 and 92TH021) resistant to neutralization by G37080 BCN plasma. (B) Neutralization breadth of the G37080 BCN plasma obtained at visit 1 and visit 2 were assessed against pseudotyped viruses expressing HIV-1 Env representing different clades and origins. Neutralization titers (median ID50 values) were obtained by titrating Env-pseudotyped viruses against G37080 plasma samples. Values at the top of each bar graph indicate the number of viruses belonging to each clade/origin tested.