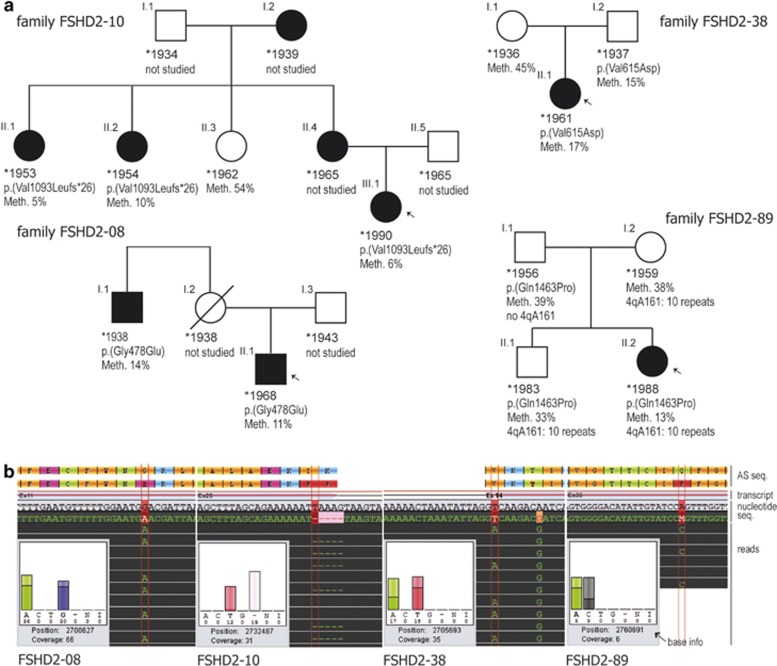
Figure 3.
Genetic characterization of four families with FSHD2. (a) Pedigrees with FSHD2. Affected individuals are depicted in black, index patients are marked with an arrow. For each individual, the following information is given if available: year of birth, variant in SMCHD1 (protein position based on NP_056110.2) and methylation level of D4Z4 in percent as determined by pyrosequencing. For family FSHD2-89, the number of repeat units on a contracted 4qA allele is given. All individuals with no indicated repeat number showed ≥11 repeat units. (b) Sequencing traces of the SMCHD1 variants from families shown above, generated by the software GenSearchNGS. The diagram shows reference and consensus sequence of protein and nucleotide, respectively (indicated as ‘AS seq.', ‘transcript' and ‘nucleotide seq.' on the right), exon/intron structure of the selected transcript (NM_015295.2) as well as base exchanges or deletions found in the displayed reads (marked by vertical red lines). The inset ‘base info' gives data on frequency and balance of the viewed base (‘A C T G' bases, ‘-' deletion, ‘N' base not defined, ‘I' insertion) and the coverage at the genomic position (hg19). In addition to the heterozygous variant, patient FSHD2-38 shows a homozygous SNP (c.1851A>G, rs635132) at g.2705700.