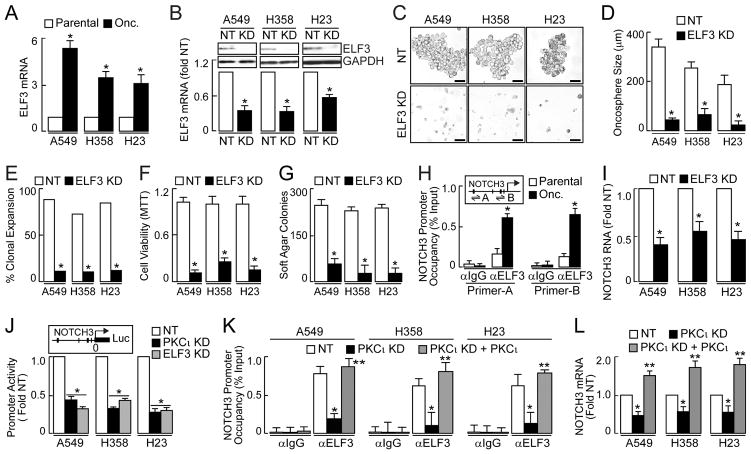
Figure 4.
Effect of PKCι on ELF3 occupancy at the NOTCH3 promoter and NOTCH3 expression. (A) QPCR for ELF3 presented as fold parental +/−SEM, n=3. *p≤ 0.05. (B) Immunoblot (top) and QPCR (bottom) of ELF3 KD. QPCR expressed as fold NT +/−SEM, n=3. *p≤ 0.05 compared to NT. (C) Representative micrographs of ELF3 KD on oncosphere growth Scale bars 50 μm. (D) Oncosphere size expressed as mean diameter (μm) +/−SEM. Oncospheres assessed: A549: NT (37), KD (37); H358: NT (34), KD (34); H23: NT (34), KD (38). *p≤0.05 compared to NT. (E) Single oncosphere cells were assessed for clonal expansion plotted as %expanded. Cells expanded/total cells scored: A549: NT (33/37), KD (5/37); H358: NT (26/34), KD (4/34); H23: NT (26/34), KD (5/38). *p≤0.001 based on Fisher’s exact test. (F) TIC viability plotted as fold NT +/−SEM, n=3, *p≤0.05 compared to NT. (G) Soft agar growth plotted as colony number +/−SEM, n=6. *p<0.05 compared to NT. (H) ChIP analysis of ELF3 occupancy of the NOTCH3 promoter in A549 TICs. Inset depicts the NOTCH3 promoter; ChIP primer-probes used are indicated as A and B. Consensus ELF3 binding sites indicated by vertical slashes. Data presented as % Input +/−SEM, n=3. *p<0.05; data representative of two independent experiments. immunoglobulin G, (IgG). (I) QPCR of NOTCH3 expressed as fold NT +/−SEM, n=3. *p≤ 0.05 compared to NT. (J) NOTCH3 promoter reporter (inset) activity plotted as fold NT +/−SEM, n=5, *p<0.05 compared to NT. (K) NOTCH3 promoter occupancy. Data presented as % Input +/−SEM, n = 3; *p<0.05. (L) QPCR for NOTCH3 expressed as fold NT +/−SEM, n=3. *p<0.05 compared to NT, **p<0.05 compared to PKCι KD. See also Figure S4.