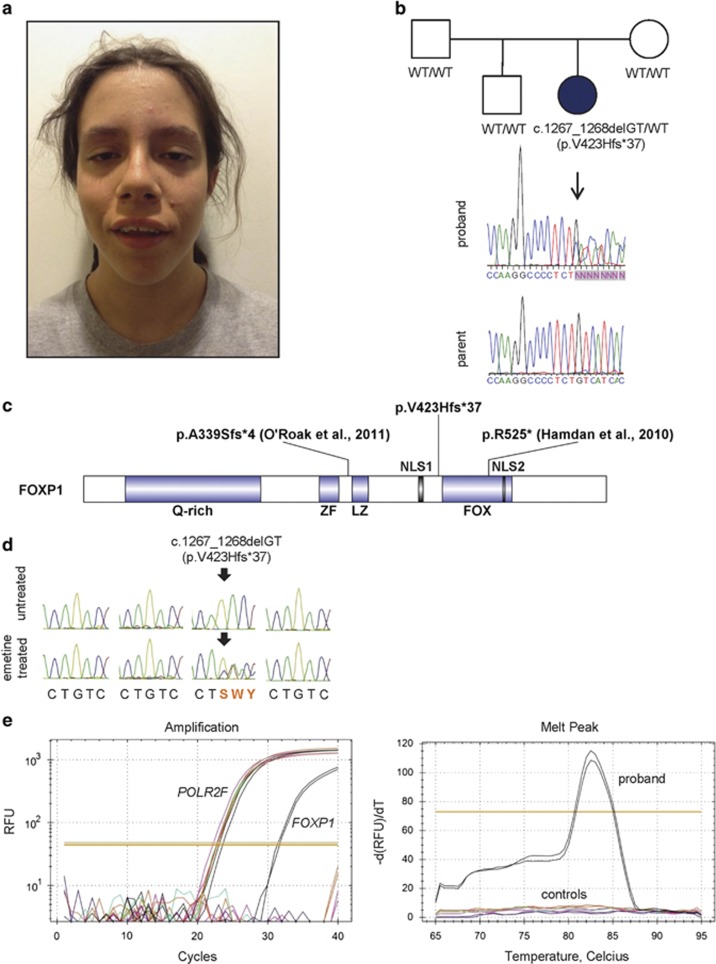
Figure 1.
Identification of a de novo FOXP1 variant in a patient with hypotonia, autism, mild ID and speech/language deficits. (a) Photograph of the patient at 14 years of age showing macrocephaly, mild ptosis, wide-spaced teeth and a hypotonic face. (b) Family pedigree showing Sanger traces of genomic DNA from the patient and one unaffected parent. The affected proband (shaded symbol) carries a heterozygous de novo two-base deletion (-GT relative to mRNA; c.1267_1268delGT) that introduces a frameshift and results in a premature stop codon. Site of deletion is indicated by black arrow. (c) Schematic representation of the FOXP1 protein indicating the change found in the patient. The FOXP1 protein has a glutamine-rich (Q-rich) region and zinc-finger (ZF), leucine zipper (LZ) and FOX DNA-binding domains, as well as two nuclear localization signals (NLS1 and NLS2). The change in FOXP1 is predicted to yield a C-terminally truncated protein (p.V423Hfs*37) that lacks the FOX DNA-binding domain and NLS. Other truncated FOXP1 variants previously reported in ASD (p.A339Sfs*4) and ID (p.R525*) are also shown. (d) Sanger traces of FOXP1 cDNA from immortalized lymphoblasts derived from the affected proband and controls (father, mother and unaffected sibling). Two sets of cultures were grown in parallel, one of which was treated with 100 μg/ml emetine for 7 h to inhibit NMD. In untreated cells, there is no evidence of the variant transcript, suggesting that the majority of these transcripts are degraded by NMD. Sanger traces from left to right: father, mother, proband, unaffected sibling. Arrows indicate site of deletion. (e) Quantitative RT-PCR amplification of variant FOXP1 transcripts in cDNA derived from the proband and controls (father, mother and unaffected sibling) (left panel). Melting curve analysis was performed to assess the specificity of the amplification (right panel). Primers were designed to specifically detect the variant allele and amplification of c.1267_1268delGT was only seen in proband-derived cDNA (black lines). Results represent two technical replicates per condition. POLR2F was used as a control. RFU, raw fluorescence unit.