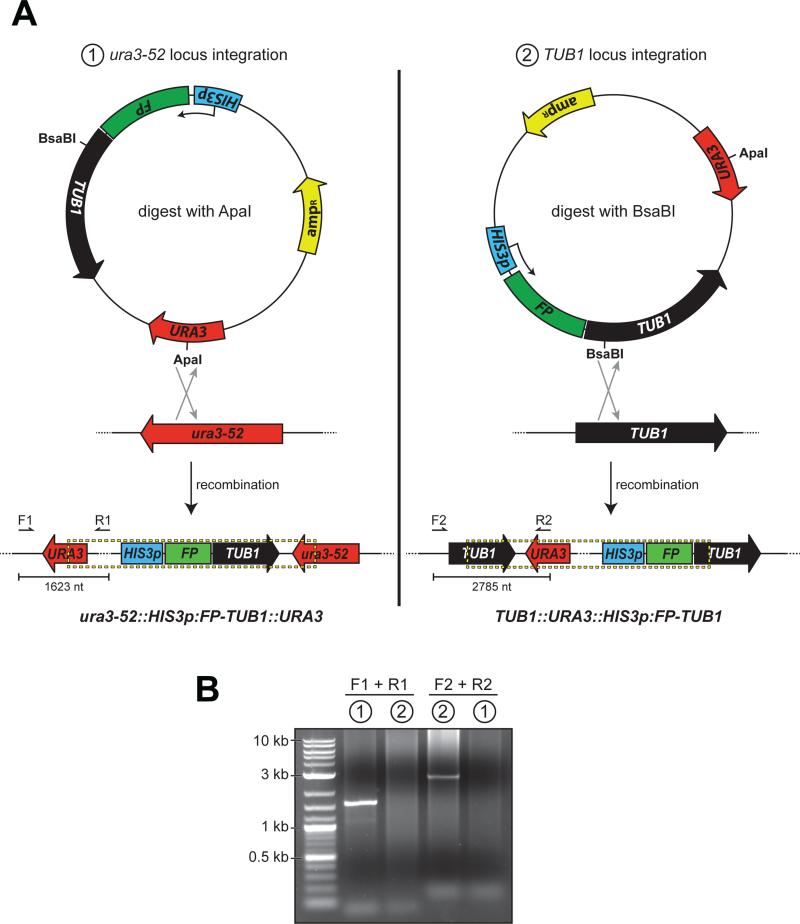
Figure 1. Differential FP-Tub1 integration strategies.
(A) Schematics depicting Tub1-tagging plasmid pRS306:HIS3p:FP-TUB1 and chromosomal loci ura3-52 and TUB1, with open reading frame and direction of coding sequence indicated (HIS3p, HIS3 promoter; FP, fluorescent protein; TUB1, alpha-tubulin; URA3, auxotrophic marker; ampR, beta-lactamase). Restriction digest by either ApaI (left; cuts within URA3 gene) or BsaBI (right; cuts within TUB1 gene) targets the plasmid for homologous recombination into either the ura3-52 or the TUB1 locus as depicted. Dashed box below delineates chromosomally-integrated plasmid. Arrows indicate the forward (i.e., F1 and F2) and reverse (i.e., R1 and R2) diagnostic PCR primers (see Table 1). (B) Ethidium bromide-stained agarose gel with PCR products from genomic DNA isolated from yeast strains transformed with either (1) ApaI-digested, or (2) BsaBI-digested pRS306:HIS3p:mCherry-TUB1 using the indicated primers. Note the specificity of each PCR product, and thus the ability to target the plasmid for chromosomal integration into either the ura3-52 or the TUB1 locus by differential restriction digest.