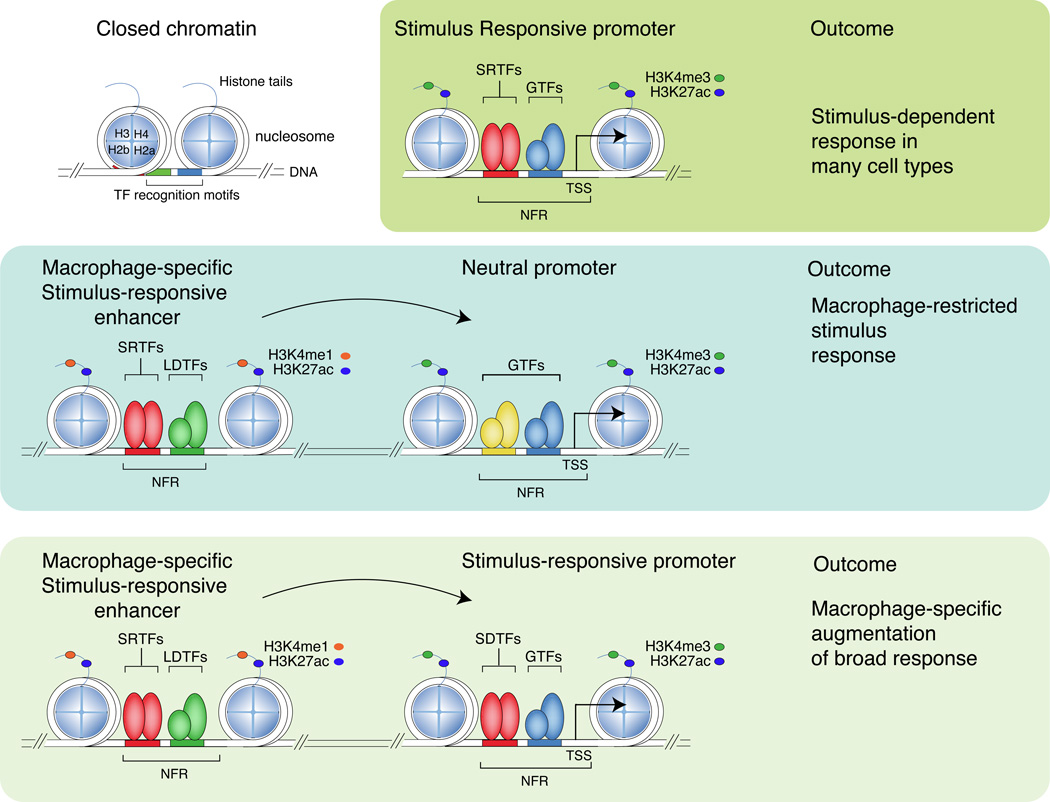
Figure 3.
Stimulus-regulated transcription factors (SRTFs) act at promoters and enhancers to direct broad or cell restricted transcriptional responses. Promoters and enhancers are primed by lineage-determining transcription factors (LDTFs) that collaborate with each other and other transcription factors to displace nucleosomes. Promoters, which are distinguished by high levels of H3K4me3 in comparison to H3K4me1, are generally primed in many cell types by broadly expressed transcription factors. Enhancers, which are distinguished by high levels of H3K4me1 in comparison to H3K4me3, are more likely to be primed by cell-type specific combinations of lineage determining transcription factors. PU.1, C/EBPs, AP-1 factors and IRFs are important macrophage lineage determining factors that drive the selection of a large fraction of macrophage-specific enhancers. Stimulus-regulated transcription factors primarily bind to DNA and activate gene expression at primed promoters and enhancers that contain their DNA recognition motifs and result in in recruitment of co-activator complexes that deposit H3K27ac. Binding of a stimulus regulated transcription factor to a promoter that is primed in many cell types is likely to result in a broad signal-dependent response. Binding of a stimulus regulated transcription factor to a cell-specific enhancer is likely to result in a cell restricted response or cell-specific potentiation of a broad response. H3K27Ac: histone H3 acetylated at lysine 27. H3K4me1: histone H3 monomethylation at lysine 4. NFR; nucleosome free region. TSS; transcriptional start site. SRTF; stimulus responsive transcription factor. GTF; general transcription factor (e.g., SP-1). LDTF; lineage determining TF (e.g. PU.1), SRTF; Stimulus responsive transcription factor, e.g., NFκB. H2a, H2b, H3, H4; histones H2a, 2b, 3 and 4, respectively.