Fig. 4.
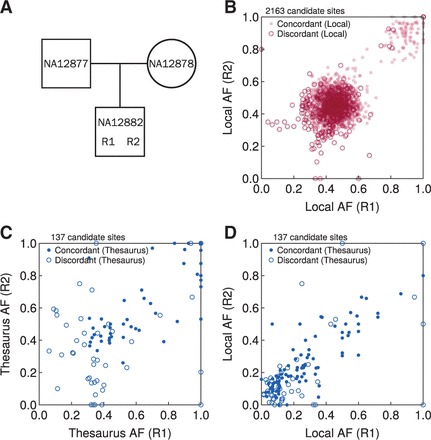
Results on de novo mutation calling from a family trio from the Platinum dataset. (A) Family pedigree for sample NA12882. R1 and R2 denote two replicates of the child’s genome. (B) Concordance of de novo mutation calls in two replicates of sample NA12882 obtained using a high (MQ 16) mapping quality threshold. Axes display the allelic frequencies (AF) of candidate sites in the two replicates. (C) Concordance of de novo mutation calls identified through thesaurus analysis of low mappability genomic regions (MQ 0) and not in the previous analysis. Allelic frequencies on the axes are informed by all thesaurus-linked genomic sites. (Thesaurus-adjusted AF > 1 are plotted at unity.) (D) Analogous to panel (C), but with allelic frequencies on the axes computed using data from only one genomic locus per mutation candidate
