Fig. 1. Assessment of errors and bias in Ig-seq using synthetic antibody spike-ins.
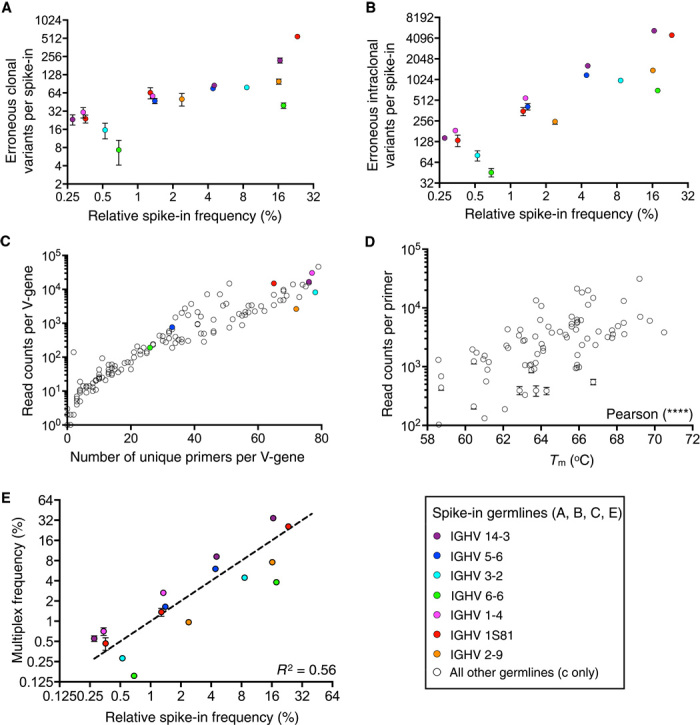
Colored dots refer to V-genes represented by spike-in clones. (A and B) Clonal (A) and intraclonal (B) diversity errors of spike-ins shown in relation to spike-in clonal frequency. (C) Mispriming of biological data during multiplex PCR is shown by plotting the number of unique primers found to be associated with a V-gene and the number of read counts per V-gene. (D) A statistically significant correlation (Pearson, two-tailed, P < 0.0001) is observed between the melting temperature (Tm) of primers in the multiplex PCR primer set and read counts associated with primers in Ig-seq data. (E) A correlation of spike-in clonal frequencies from library preparation with multiplex PCR versus singleplex PCR results in an R2 = 0.56. Amplification bias was systematic because error bars were very low across replicate sequencing runs. Clones with the same V-gene were consistently under- or overamplified. Ig-seq data are from replicate library sample preparations (n = 3; data sets consisted of 4 × 105 preprocessed full-length antibody reads) from mouse splenic cDNA with synthetic spike-ins [for (A), (B), (D), and (E), data are presented as means ± SD and are from replicate data sets Reddy-PS-1, Reddy-PS-2, and Reddy-PS-3; data set Reddy-PS-1 was used for (C); see table S2]. Relative spike-in frequencies are mean values obtained from replicate libraries (n = 5) generated by singleplex PCR (see fig. S2 and table S1).
