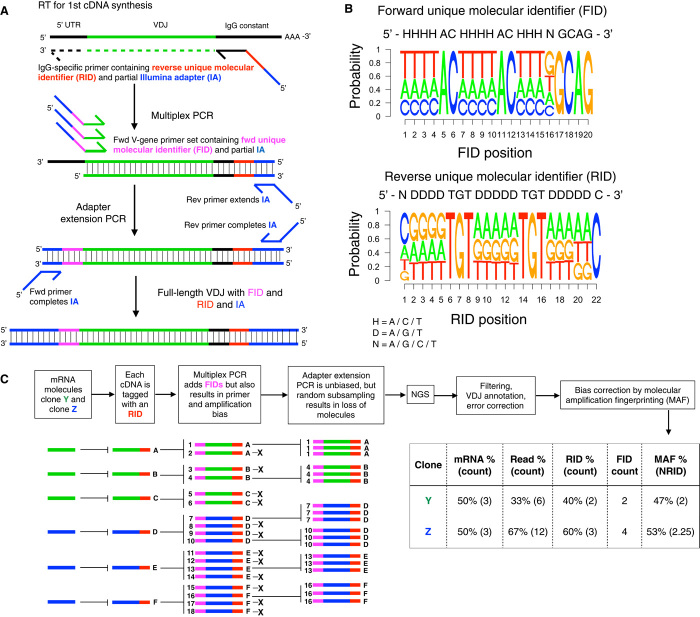
Fig. 2. Ig-seq with MAF.
(A) Workflow for library preparation by MAF consists of reverse transcription (RT), multiplex PCR, and adapter extension PCR and results in amplicons ready for Ig-seq. (B) Following Ig-seq, nucleotide sequence logos show FID and RID regions with predicted levels of variability and nonvariability in degenerate and spacer regions, respectively. (C) Schematic shows the principle of MAF bias correction and its ability to provide improved accuracy of clonal frequencies. The MAF % is based on the normalized RIDcount (NRID), which is equal to the RID clonal counts divided by the MAF bias factor (FIDclonal count/RIDclonal count). In the example above, the NRID for clone Y = (2)/(2/2) = 2; clone X = 3/(4/3) = 2.25 (see Results for more details).