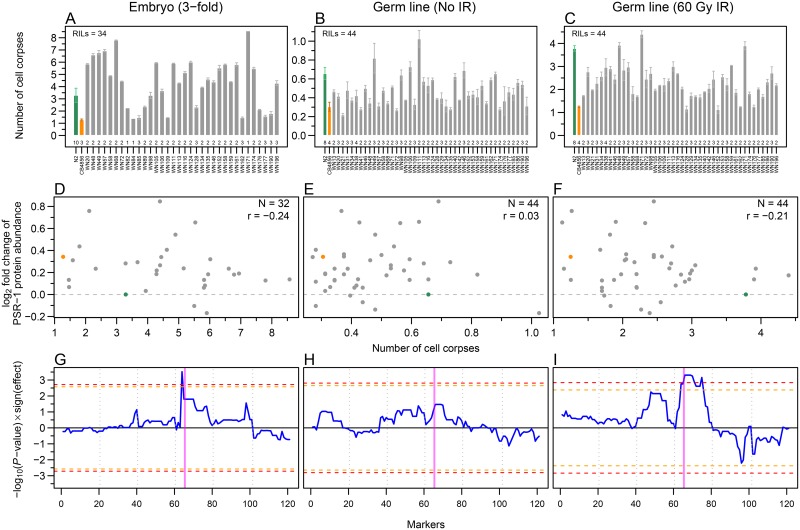
Fig 7. Analysis of variation in embryonic and germ line apoptosis in parental strains and RILs.
(A-C) Quantification of apoptotic cell corpse numbers in embryos (A), germ line without ionizing radiation (IR; B), and with 60 Gy IR (C). Error bars represent SEM between numbers of biological replicates indicated at the bottom of each bar. (D-F) Natural variation in apoptosis levels of parental strains and RILs does not correlate with the PSR-1 protein abundance (relative to N2). Scatter plots (CB4856 in orange and N2 in green) of PSR-1 protein abundance and apoptotic levels in embryos (D), germ line without IR (E), and with 60 Gy IR (F). Pearson correlation coefficient is denoted by r. (G-I) Embryonic and IR-induced apoptosis shows a significant QTL on chromosome IV. QTL profiles of apoptotic phenotype in embryos (G), adult germ line without IR (H) and with 60 Gy IR (I). Blue curves show the significance of the QTLs multiplied by the sign of the effect of the N2 allele (positive values of blue curve indicate higher apoptosis level when the N2 allele is present, whereas negative values indicate higher apoptosis level when the CB4856 allele is present). Horizontal orange and red dashed lines show 0.1 and 0.05 FDR thresholds respectively. Vertical dotted grey lines separate chromosomes I to V and X from left to right. Vertical magenta band indicates the position of psr-1 gene in the genome.