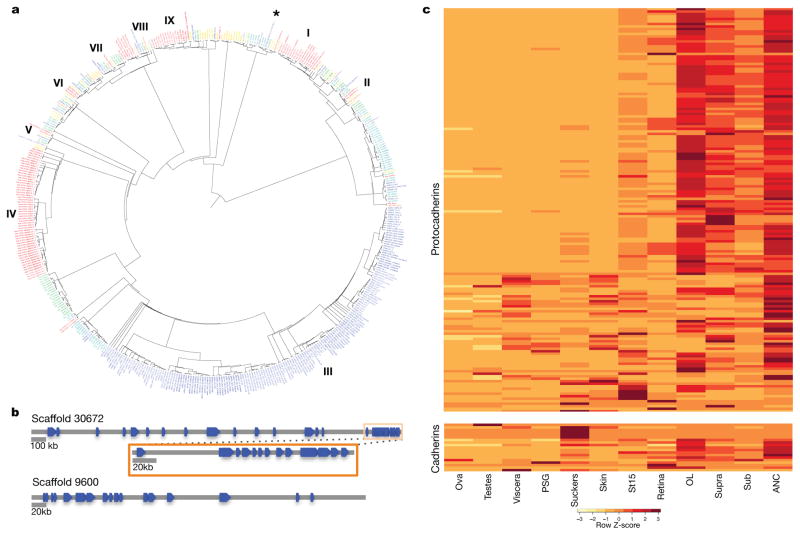
Figure 2. Protocadherin expansion in octopus.
a, Phylogenetic tree of cadherin genes in Hsa (red), Dme (orange), Nematostella vectensis (Nve, mustard yellow), Amphimedon queenslandica (Aqu, yellow), Cte (green), Lgi (teal), Obi (blue), and Saccoglossus kowalevskii (Sko, purple). I, Type I classical cadherins; II, calsyntenins; III, octopus protocadherin expansion (168 genes); IV, human protocadherin expansion (58 genes); V, dachsous; VI, fat-like; VII, fat; VIII, CELSR; IX, Type II classical cadherins. Asterisk denotes a novel cadherin with over 80 extracellular cadherin domains found in Obi and Cte. b, Scaffold 30672 and Scaffold 9600 contain the two largest clusters of protocadherins, with 31 and 17, respectively. Clustered protocadherins vary greatly in genomic span and are oriented in head-to-tail fashion along each scaffold. c, Expression profiles of 161 protocadherins and 19 cadherins in 12 octopus tissues; 7 protocadherins were not detected in the tissues sampled. Cells are colored according to number of standard deviations from the mean expression level. Protocadherins have high expression in neural tissues. Cadherins generally show a similar expression pattern, with the exception of a group of sucker-specific cadherins.