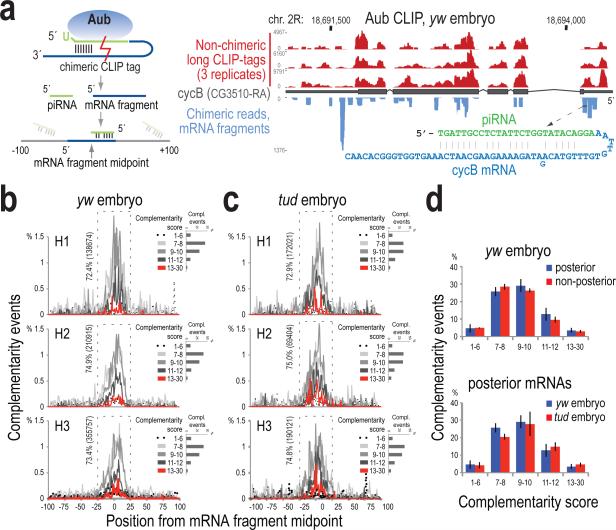
Figure 2. Complementarity analysis between the piRNA and mRNA parts of chimeric CLIP tags.
a. Strategy for chimeric CLIP tag analysis, and genome browser illustrating Aub lgClips on cycB; sequence and base pairing of a chimeric CLIP tag is shown.
b, c. piRNA:mRNA complementarity events (percent) within ±100 bases from the midpoint of the mRNA part of the chimeric read, plotted per alignment score for yw (b) and tud embryo (c) Aub CLIPs (biological triplicates). Percentage and number of total events occurring within ±25 bases (dashed rectangle) are shown. Inset, per sample: barplot of number of complementarity events per score group.
d. Barplots of piRNA:mRNA complementarity events occurring within the ±25 bases window of the midpoint of the search area and with score ≥7, for indicated mRNA localization categories and Aub CLIP libraries. Error bars: ±S.D.; n=3.