Abstract
Genes have preferential non-random spatial positions within the cell nucleus. The nuclear position of genes may differ between cell types and some genes undergo repositioning events in disease, including cancers. It is currently unclear whether the propensity of a gene to reposition reflects an intrinsic property of the locus or the tissue. Using quantitative FISH analysis of a set of genes which reposition in cancer, we test here the tissue-specificity of gene repositioning in normal and malignant breast or prostate tissues. We find tissue-specific organization of the genome in normal breast and prostate tissues with 40% of genes occupying differential positions between the two tissue types. While we demonstrate limited overlap between gene sets that repositioned in breast and prostate cancer, we identify two genes that undergo disease-related gene repositioning in both cancer types. Our findings indicate that gene repositioning in cancer is tissue-of-origin specific.
Keywords: Nuclear architecture, spatial gene positioning, breast cancer, prostate cancer, tissue specificity
Introduction
The genome is non-randomly organized within the interphase nucleus (Ferrai et al. 2010; Meaburn et al. 2016). There is a general tendency for gene rich and gene poor genomic regions, ranging from whole chromosomes to chromatin domains, to be spatially separate from each other (Boutanaev et al. 2005; Boyle et al. 2001; Croft et al. 1999; Guelen et al. 2008; Lieberman-Aiden et al. 2009; Shopland et al. 2006). While this trend is maintained across species and between cell types, the preferred nuclear locations of specific chromosomes and genes can be variable depending on cellular context. Spatial reorganization of the genome occurs, for example, with changes in proliferation status, during differentiation, between cell types and in disease (Ferrai et al. 2010; Meaburn et al. 2016). It is currently unclear how the spatial arrangements of the genome are established and maintained, although epigenetic modifications, chromatin remodeling, gene expression and replication timing have been implicated for the positioning of some genes (Ferrai et al. 2010; Hiratani et al. 2008; Kosak et al. 2002; Meaburn et al. 2016; Peric-Hupkes et al. 2010; Therizols et al. 2014; Towbin et al. 2012; Volpi et al. 2000; Williams et al. 2002).
The organization of the genome between terminally differentiated cell types is remarkably similar despite profound differences in function, gene expression profiles and nuclear shape. For instance, most human chromosomes (HSAs) occupy similar radial positions in lymphoblast and dermal fibroblast nuclei, with only HSAs 8, 20 and 21 in different positions (Boyle et al. 2001; Meaburn et al. 2008). This phenomenon is not limited to humans, as there is also a high level of conservation in chromosomal positioning patterns between porcine lymphoblasts and fibroblasts, where chromosome 17 is the only differentially positioned chromosome (Foster et al. 2012). In mice, three of six chromosomes analyzed position similarly between lymphoblasts and fibroblasts (Mayer et al. 2005). Comparisons of positioning patterns of chromosomes between other cell types are less extensive; however, there is conservation in positioning patterns for at least some chromosomes. The positioning of HSA 18 and HSA 19 is broadly maintained in a wide range of cell types, including fibroblasts, keratinocytes, lymphocytes and epithelial cells from multiple tissues (Boyle et al. 2001; Cremer et al. 2003; Murata et al. 2007). Moreover, porcine chromosome 17, 13, 5 and X are similarly positioned between kidney, lung and brain tissue (Foster et al. 2012) and mouse chromosome (MMU) 14 is similarly positioned in lung and kidney cells (Parada et al. 2004). However, tissue-specific positioning of specific chromosomes does also occur. All six mouse chromosomes analyzed in freshly isolated small and large lung cells, liver, kidney, lymphocytes and myeloblasts cells are in differential positions in at least three tissue types (Parada et al. 2004). For example, MMU 12 and 15 are in distinct positions in lung and kidney cells, yet are in similar positions in lymphocytes and myeloblasts (Parada et al. 2004). Moreover, MMU 5 was the only analyzed chromosome in a different spatial position between lymphocytes and myeloblasts, whereas MMU 1, 5, 6 and 12 are differentially positioned between liver and myeloblasts (Parada et al. 2004). These observations point to partial conservation of positioning patterns amongst tissues.
Similar yet distinct spatial arrangement of the genome between differentiated cell types is also consistent with genome-wide analyses (Battulin et al. 2015; Peric-Hupkes et al. 2010). Approximately 80% of genomic regions that associate with the nuclear lamina protein lamin B1, used as a marker of the nuclear periphery, in in vitro differentiated mouse astrocytes also preferentially associate with the nuclear lamina in embryonic fibroblasts (Peric-Hupkes et al. 2010). Moreover, despite the highly compact and haploid nature of DNA in sperm, the genome-wide spatial organization of DNA is similar in mouse sperm and fibroblast nuclei when analyzed by Hi-C genome-wide crosslinking methods (Battulin et al. 2015). The organization is, however, not fully identical. For example, there is a higher frequency of long-range interactions in sperm compared to fibroblasts (Battulin et al. 2015). Additionally, in other cell types, some gene loci have tissue-specific positions. For example, human CFTR is more peripherally positioned in lymphocytes and embryonic kidney cells compared to nasal epithelial cells and CORTBP2 is more peripherally positioned in embryonic kidney cells compared to lymphocytes (Zink et al. 2004). Furthermore, the spatial position of a gene with respect to the chromosome it resides on may also vary between cell types. Some gene rich clusters of functionally related genes, such as the epidermal differentiation complex or the major histocompatibility complex, loop out from the bulk of their chromosome territory more frequently in cell types where they are highly expressed (Volpi et al. 2000; Williams et al. 2002). Other genes, such as PAX6, WT1, DMD, FLNA and BCL2, however, remain within the chromosome territory in different cell types, independent of transcription status (Kurz et al. 1996; Mahy et al. 2002; Scheuermann et al. 2004).
Spatial reorganization of the genome is also associated with disease (Borden and Manuelidis 1988; Meaburn et al. 2007; Mehta et al. 2011; Mewborn et al. 2010; Mikelsaar et al. 2014; Paz et al. 2015), including cancer (Cremer et al. 2003; Leshner et al. 2015; Meaburn et al. 2009; Meaburn and Misteli 2008; Murata et al. 2007; Wiech et al. 2009; Zeitz et al. 2013). Much like between tissue types, changes in genome organization are not global and only subsets of gene loci alter their nuclear location (Borden and Manuelidis 1988; Leshner et al. 2015; Meaburn et al. 2007; Meaburn et al. 2009; Meaburn and Misteli 2008; Zeitz et al. 2013). We have previously identified two sets of genes that repositioned in malignant breast or prostate tissue, respectively, compared to their normal counterparts (Leshner et al. 2015; Meaburn et al. 2009). These repositioning events were specific to the malignant state and do not commonly occur in non-malignant disease and they are not accounted for by inter-individual variations, nor do they correlate with numerical genome abnormalities, gene ontology, the local gene density surrounding the loci or changes in gene expression (Leshner et al 2015; Meaburn et al. 2009; Meaburn and Misteli 2008). In line with partial repositioning of genomes in cancer, the majority of genes did not change radial positioning in neither breast nor prostate cancer, suggesting gene-specific repositioning events (Leshner et al. 2015; Meaburn et al. 2009; Meaburn and Misteli 2008).
A key question that emerges from these observations is whether the same genes reposition in multiple cancers or whether the set of repositioned genes is tissue-specific. Previous analysis has found repositioning of HSA 18 and 19 in many types of cancer, including Hodgkin’s lymphoma, melanoma, colon, cervical and thyroid carcinomas, suggesting a lack of tissue-of-origin specificity (Cremer et al. 2003; Murata et al. 2007; Wiech et al. 2009). In contrast to the other types of cancer, however, HSA 18 is more peripheral in a cervical squamous carcinoma tissue (Wiech et al. 2009). Moreover, BCL2 is relocated to a more peripheral nuclear position in a BCL2 expressing cervical squamous carcinoma (Wiech et al. 2009), but its position is unaffected in breast cancer (Meaburn et al. 2009), prostate cancer (Leshner et al. 2015) and in a BCL2 negative cervical squamous carcinoma (Wiech et al. 2009), pointing to tissue-specific differences in repositioning behavior.
To more systematically determine whether cancer-associated gene repositioning is gene- or tissue-of-origin specific, we have compared here the nuclear positions of a set of eleven genes, which we have previous identified to robustly reposition in either breast or prostate cancers. We find that the repositioned genes are largely distinct in each tissue type with only two genes repositioning in both types of cancer. These results point to tissue-of-origin specificity for gene repositioning in cancer.
Materials and Methods
Tissue FISH
To generate fluorescence in situ hybridization (FISH) probes, bacterial artificial chromosome (BAC) clones (BACPAC resource center) (Suppl. Table S1) were label with either biotin- or digoxigenin-conjugated dUTPs (Roche) by nick translation, as previously described (Meaburn 2010; Meaburn et al. 2009). FISH was performed on 4–5μm thick FFPE de-identified human tissue sections (Suppl. Table S2), as previously described in (Meaburn et al. 2009) and using the following modifications: the 60°C slide baking step was not performed, tissue sections were incubated in 0.25 mg/ml Proteinase K (Sigma-Aldrich) for 15–20 mins, except for single tissue slides from Biomax Inc, where 0.5mg/ml Proteinase K was typically required. Tissue sections and tissue microarrays (TMAs) were purchased from US Biomax Inc, Imgenex Corporation, Folio Bioscience and Biochain Institute or were acquired from the University of Washington under the guidelines and approval of the Institutional Review Board of the University of Washington (# 00-3449) (Suppl. Table S2). The prostate tissues from the University of Washington were reviewed by a genitourinary pathologist (L.D.T.). The panel of tissues included twelve breast cancers, six benign breast tissues (hyperplasia and fibroadenoma), six normal breast tissues, 20 prostate cancers, four hyperplasic prostate tissues and 24 histologically benign (normal) prostate specimens (Suppl. Table S2).
Image acquisition and FISH analysis
Image accusation was performed as previously described (Leshner et al. 2015; Meaburn et al. 2009). Briefly, all imaging was performed on a wide-field IX70 (Olympus) Deltavision (Applied Precision) microscope system, equipped with a 60× 1.42N oil objective lens (Olympus). An auxiliary magnification of 1.5 and a z-step size of 0.5μm were used to acquire image stacks. Most tissue sections or TMA tissue cores contained predominantly a single morphology (e.g. malignant tissue only), and regions of epithelial nuclei were randomly imaged over the slide or tissue core. The prostate tissues from the University of Washington, however, often contained multiple morphologies on the same slide. In these cases, malignant and non-malignant glands were imaged and analyzed separately, after examination of the tissue at low resolution (10× lens; Olympus) and consultation of hematoxylin and eosin stained slides, which had been annotated by a pathologist (L.D.T.). Images of epithelial nuclei where then randomly acquired within the predetermined regions of benign or malignant tissue. Before image analysis, 2D maximum intensity projections were generated from deconvolved image stacks, using SoftWoRx (Applied Precision) as previously described (Meaburn et al. 2009).
Analysis to map the spatial position of FISH signals was performed as previously described (Leshner et al. 2015; Meaburn et al. 2009). Briefly, 89 to 169 interphase epithelial nuclei, which had been manually segmented in Photoshop 7.0 (Adobe) based on DAPI staining, were run though custom image analysis algorithms (Meaburn et al. 2009), which were executed in MATLAB (The Mathsworks Inc.) and utilized DIPImage and PRTools toolboxes (Deft University). To determine the radial position of each FISH signal, each pixel of a nucleus was labeled with the distance to the nearest nuclear boundary, by Euclidean distance transform (EDT), to enable the nuclear EDT value of the geometric gravity center of each FISH signal to be automatically determined. Next, the EDT value for a FISH signal was normalized to the maximal nuclear EDT for that nucleus, to normalize for variations in nuclear size. To generate a relative radial distribution (RRD) for a gene in a given specimen, the normalized FISH signal EDT for every allele from that tissue was combined and a cumulative frequency distribution was produced. For each gene a PND was also created. To this end, the normalized EDTs from all allele in each normal tissue, for a given gene, were combined. Table 1 details the number of normal tissues analyzed for each gene and Suppl. Table S1 details the number of nuclei used to produce each PND. Finally, to statistically compare a gene’s positioning patterns between individuals and between histological states, cumulative RRDs between tissues were cross-compared using the nonparametric two-sample 1D Kolmogorov-Smirnov (KS) test, with P < 0.01 considered significant.
Table 1.
Comparison of gene positioning patterns between normal breast and normal prostate tissues
| Gene | % (and number) of SD cross-comparison between individual normal breast and prostate tissues | Tissue specific positioning?a |
|---|---|---|
| AKT1 | 39.3% (11/28)b | No |
| CSF1R | 64.9% (50/77)b | Yes |
| ERBB2 | 30.6% (11/36)b | No |
| FLI1 | 20.8% (10/48)c | No |
| FOSL2 | 46.0% (29/63)b | No |
| HES5 | 14.3% (6/42)b | No |
| HSP90AA1 | 61.2% (26/42)b | Yes |
| MMP2 | 28.1% (9/32)c | No |
| MMP9 | 86.7% (52/60)c | Yes |
| MYC | 97.2% (35/36)b | Yes |
| TGFB3 | 25.0% (12/48)b | No |
|
| ||
| BCL2 | 100.0% (12/12)b,c | Yes |
| CCND1 | 37.5% (6/16)b,c | No |
| MMP1 | 66.7% (4/6)b,c | Yes |
| VEGF | 9.6% (5/52)b,c | No |
SD, significantly different, based on a two-sample 1D KS test, P < 0.01.
between normal breast and prostate tissue. These comparisons utilize positioning RRDs (position distributions) previously generated in:
RRDs from breast tissues were previously published in (Meaburn et al. 2009) or
prostate tissue RRDs from (Leshner et al. 2015).
Some previously reported data were included in the current analysis for comparison of the positioning pattern of a gene between normal breast and prostate tissues. The RRDs for AKT1, CSF1R, ERBB2, FOSL2, HES5, HSP90AA1, MYC, TGFB3, BCL2, CCND1, MMP1 and VEGFA in normal breast tissues were reported in (Meaburn et al. 2009) and the RRDs for FLI1, MMP9, MMP9, BCL2, CCND1, MMP1 and VEGF in prostate tissue are as reported in (Leshner et al., 2015).
Results
We have previously identified a set of eight genes (AKT1, CSF1R, ERBB2, FOL2, HES5, HSP90AA1, MYC and TGFB3) that reposition in breast cancer (Meaburn et al. 2009) and a set of three genes (FLI1, MMP2 and MMP9) that reposition in prostate cancer (Leshner et al. 2015). We refer to these gene sets as breast GBPs (Gene Positioning Biomarkers) and prostate GPBs, respectively. To determine whether the observed repositioning events between normal and cancer tissues are tissue-specific, we sought to cross-compare prostate GPBs and breast GPBs in their heterologous tissues. To this end, we performed FISH to visualize the GPBs (Suppl. Table S1) in 4–5μm thick formalin-fixed, paraffin embedded (FFPE) human tissues (Suppl. Table S2). We positioned prostate GPBs in benign (normal, hyperplasia, fibroadenoma) and malignant breast tissues and, conversely, breast GPBs in normal, normal adjacent to tumor (NAT) and adenocarcinoma prostate tissues (Suppl. Table S2). Typically 100–150 randomly selected epithelial nuclei were analyzed per tissue for each gene as previously described (Leshner et al. 2015; Meaburn et al. 2009). The radial position of each allele, normalized to nuclear size, was determined from projections of image stacks using EDT, as previously described (see Materials and Methods for details; Leshner et al. 2015; Meaburn et al. 2009). The normalized positions for each allele in a tissue were then combined to generate a cumulative RRD for each gene. Statistical differences between samples were assessed using the two-sample 1-D KS test, as described (see Materials and Methods; Leshner et al. 2015; Meaburn et al. 2009), and P < 0.01 was considered significant.
Tissue-specific spatial organization in normal tissues
To first determine tissue-specificity of gene positioning in normal tissues, we compared the positioning patterns of the full set of eleven genes between multiple normal breast and prostate tissues (Table 1, Suppl. Tables S2–S3, Fig. 1a, b, Suppl. Fig. S1a). To increase the number of genes compared between the tissue types, for this analysis we included three additional genes that do not reposition significantly in breast or prostate cancer (BCL2, CCND1 and MMP1) and VEGF, which repositioned in approximately half of both prostate and breast cancers (Table 1, Suppl. Table S4, Fig. 1c, Suppl. Fig. S1b). Many genes exhibited significantly overlapping distributions between normal breast and prostate tissues. For six genes (40%; VEGFA, HES5, FLI1, TGFB3, MMP2 and ERBB2) less than a third of the pair-wise cross-comparisons of RRDs between normal breast and prostate tissue were significantly different and for an additional three genes (20%; CCND1, AKT1 and FOSL2) 37.5–46.0% of the cross-comparisons between the tissue types were significant (Table 1, Fig. 1, Suppl. Fig. S1). On the other hand, six of the 15 genes (40%; HSP90AA1, CSF1R, MMP1, MMP9, MYC, and BCL2) displayed distinct tissue-specific positioning, with 61.2–100% of the pairwise comparisons between normal breast and prostate tissue being significantly different (Table 1, Fig. 1, Suppl. Fig. S1). These observations point to a partial conservation of gene positioning between breast and prostate tissues.
Fig. 1.
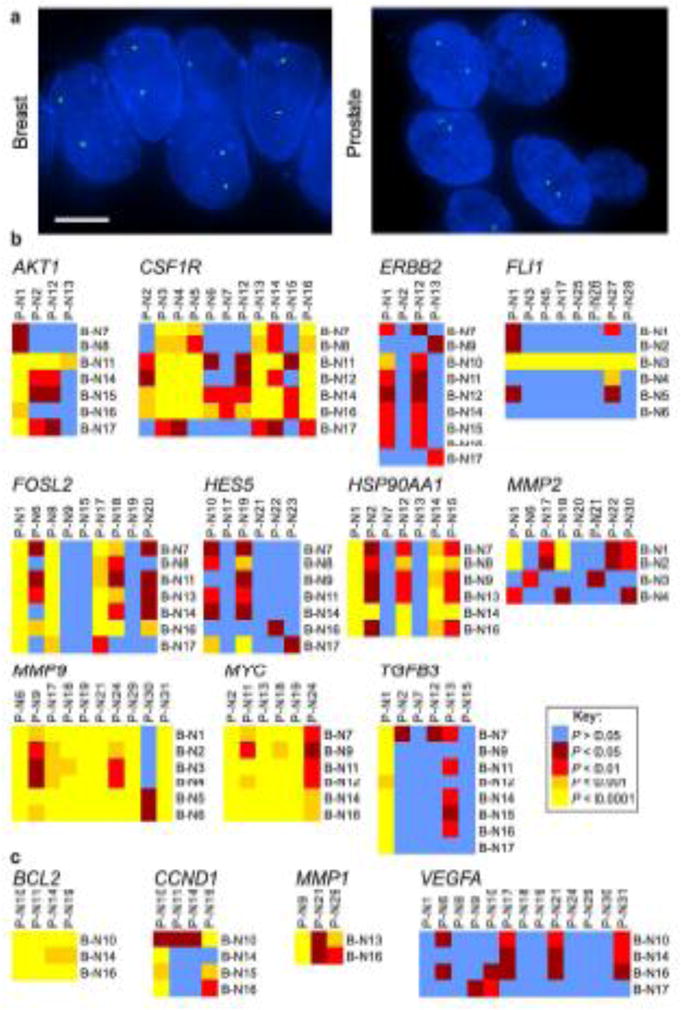
Tissue-specific spatial organization of the genome. (a) FISH was used to detect gene loci in FFPE tissue sections. FLI1 (green) in a normal breast tissue and HES5 (green) in a normal prostate tissue. Blue, DAPI nuclear counterstain. Scale bar, 5μm. Projected image stacks are shown. (b, c) Heat maps representing the pair-wise statistical comparisons of the positioning patterns of the indicated genes between normal breast tissues (B-N1 – B-N17; see Suppl. Tables S2, S3) and normal prostate tissues (P-N1 – P-N31; see Suppl. Tables S2, S3), using the two-sample 1D KS test. (b) Genes that reposition in either breast or prostate cancer (or both). (c) Genes that do not significantly reposition in either breast or prostate cancer. For these comparisons, the RRDs for AKT1, CSF1R, ERBB2, FOSL2, HES5, HSP90AA1, MYC, TGFB3, BCL2, CCND1, MMP1 and VEGFA in normal breast tissues are from (Meaburn et al. 2009) and the RRDs for FLI1, MMP9, MMP2, BCL2, CCND1, MMP1 and VEGF in prostate tissue are from (Leshner et al. 2015)
Distinct sub-sets of genes reposition in breast and prostate cancer
To address if similar repositioning events occur in multiple cancer types or if distinct sets of genes reposition in cancers originating from different tissues, we compared the repositioning behavior of breast GPBs in prostate cancer and vice versa. To this end, we first compared the RRDs of the prostate GPBs, FLI1, MMP9 and MMP2, in malignant breast tissues to individual normal breast tissues and to a pooled normal distribution (PND; Tables 2–3, Figs. 2–3), which was generated by averaging the RRDs from all normal tissues analyzed into a single distribution for each gene, as previously described (Leshner et al. 2015; Meaburn et al. 2009; see Materials and Methods). We find that MMP2 was similarly positioned in breast cancer and normal breast tissue with 37.5% (9/24) of individual cross-comparisons between normal and malignant breast tissues significantly different. In addition, the distribution of MMP2 was not statistically significantly different from the PND in all six breast cancer tissues (Tables 2, 3, Figs. 2b–3). Furthermore, the position of MMP2 was more different amongst normal tissues than when normal and cancer tissue was compared (Tables 2–3, Figs. 2b–3). In contrast to MMP2, the prostate GPBs FLI1 and MMP9 also repositioned in breast cancer (Tables 2–3, Figs. 2–3). For both genes, 66.7% (40/60 and 48/72, respectively) of cross-comparisons between normal breast and breast cancer were significantly different. Moreover, compared to the PND, FLI1 was repositioned in all ten breast cancers and MMP9 was repositioned in 83.3% (10/12) of breast cancers. Importantly, FLI1 and MMP9 were both similarly positioned amongst normal breast tissues, with only 33.3% (5/15) and 6.7% (1/15), respectively, of individual cross-comparisons between normal breast tissues reaching significance. Finally, compared to their PNDs, FLI1 was repositioned in a single (1/6) normal tissue and MMP9 was repositioned in none (0/6) of the normal breast tissues (Tables 2–3, Figs. 2b–3).
Table 2.
Cross-comparisons between individual tissues
| Gene | Tissue | Number of tissues | % (and number) of SD cross-comparison between: | ||
|---|---|---|---|---|---|
| Normal | Cancer | Individual normal tissues | Individual normal and cancer tissues | ||
| FLI1 | Breast | 6 | 10 | 33.3% (5/15) | 66.7% (40/60) |
| MMP9 | Breast | 6 | 12 | 6.7% (1/15) | 66.7% (48/72) |
| MMP2 | Breast | 4 | 6 | 50.0% (3/6) | 37.5% (9/24) |
|
| |||||
| AKT1 | Prostate | 4 | 6 | 16.7% (1/6) | 37.5% (9/24) |
| CSF1R | Prostate | 11 | 9 | 41.8% (23/55) | 38.4% (38/99) |
| ERBB2 | Prostate | 4 | 5 | 33.3% (2/6) | 35.0% (7/20) |
| FOSL2 | Prostate | 9 | 7 | 38.9% (14/36) | 47.6% (30/63) |
| HES5 | Prostate | 6 | 6 | 20.0% (3/15) | 27.8% (10/36) |
| HSP90AA1 | Prostate | 7 | 4 | 57.1% (12/21) | 46.4% (13/28) |
| MYC | Prostate | 6 | 5 | 6.7% (1/15) | 3.3% (1/30) |
| TGFB3 | Prostate | 6 | 4 | 33.3% (5/15) | 25.0% (6/24) |
SD, significantly different, based on a two-sample 1D KS test, P < 0.01.
Table 3.
Comparison of individual tissues to a pooled normal distribution
| Gene | Tissue | % (and number) of SD cross-comparison between: | |
|---|---|---|---|
| Individual normal tissues and pooled normal | Individual cancer tissues and pooled normal | ||
| FLI1 | Breast | 16.7% (1/6) | 100.0% (10/10) |
| MMP9 | Breast | 0.0% (0/6) | 83.3% (10/12) |
| MMP2 | Breast | 25.0% (1/4) | 0.0% (0/6) |
|
| |||
| AKT1 | Prostate | 0.0% (0/4) | 50.0% (3/6) |
| CSF1R | Prostate | 27.3% (3/11) | 44.4% (4/9) |
| ERBB2 | Prostate | 0.0% (0/4) | 40.0% (2/5) |
| FOSL2 | Prostate | 33.3% (3/9) | 42.9% (3/7) |
| HES5 | Prostate | 0.0% (0/6) | 33.3% (2/6) |
| HSP90AA1 | Prostate | 42.9% (3/7) | 50.0% (2/4) |
| MYC | Prostate | 0.0% (0/6) | 0.0% (0/5) |
| TGFB3 | Prostate | 16.7% (1/6) | 25.0% (1/4) |
SD, significantly different, based on a two-sample 1D KS test, P < 0.01.
Fig. 2.
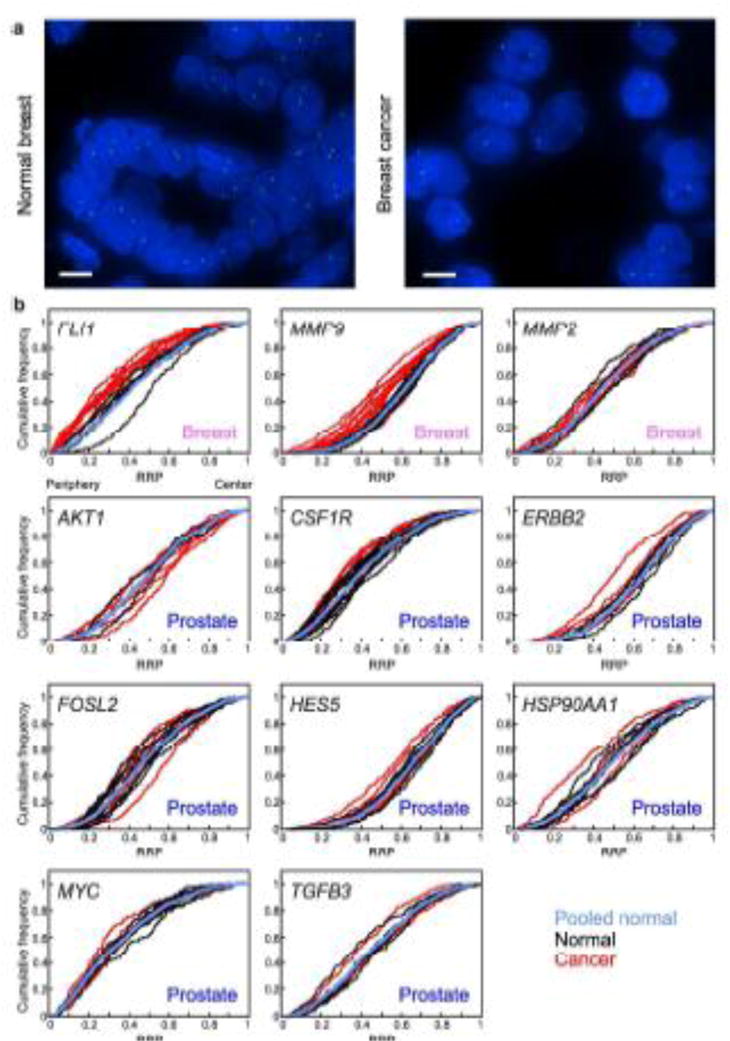
Positioning patterns in cancer compared to normal tissue. (a) MMP9 (green) was detected by FISH in normal and cancerous breast FFPE tissue sections. Blue, DAPI nuclear counterstain. Scale bar, 5μm. Projected image stacks are shown. (b) Cumulative RRDs for the indicated genes in cancer (red), normal tissues (black) and the pooled normal distribution (blue). The tissue type is specified in each graph. RRP, relative radial position
Fig. 3.
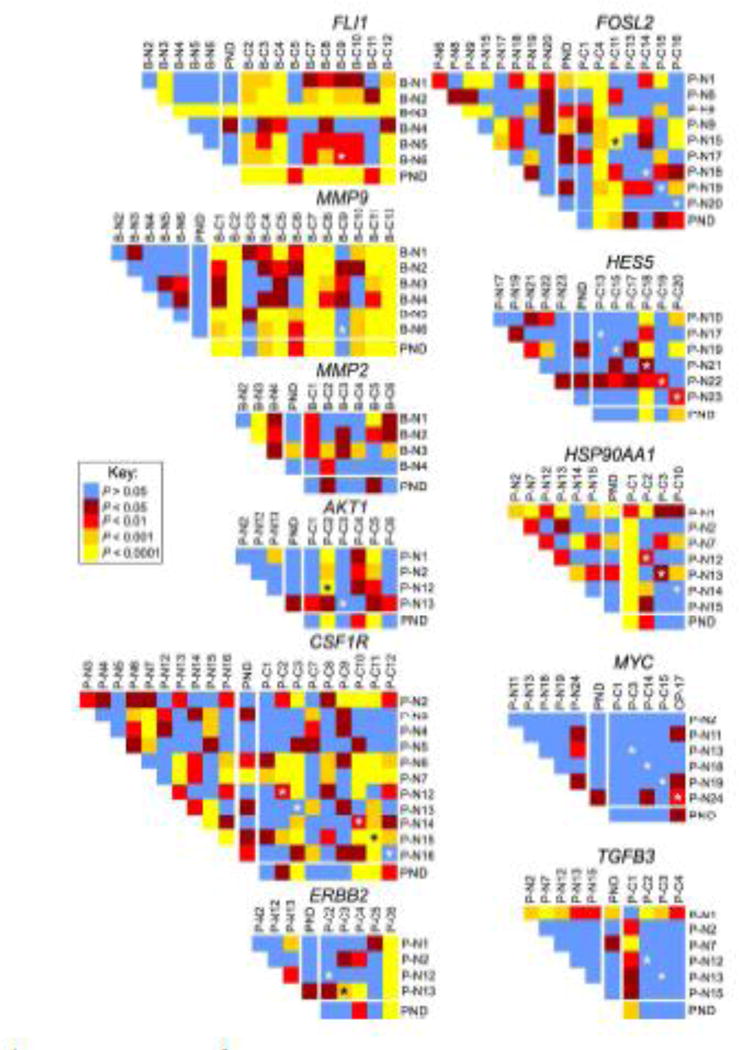
Statistical cross-comparisons of gene positioning in cancer tissues. Heat maps representing the pair-wise statistical comparisons of positioning patterns of indicated genes between tissues, using the two-sample 1D KS test. B-N1 – B-N6, normal breast tissues (see Suppl. Table S2); B-C1 – B-C12, cancerous breast tissues; P-N1 – P-N24, normal prostate tissues; P-C1 – P-C20, cancerous prostate tissues; PND, pooled normal distribution. Black or white asterisks indicate a cross-comparison between a normal and cancer specimen from the same individual
In an analogous fashion, we next determined how many of the previously identified breast GPBs (AKT1, CSF1R, ERBB2, FOL2, HES5, HSP90AA1, MYC and TGFB3) reposition in prostate cancer. We find that none of these breast GPBs robustly reposition in prostate cancer tissue (Tables 2b–3, Figs. 2–3). The majority of cross-comparisons between normal and prostate cancer tissues were not significantly different for any of the genes with between 3.3%–47.6% cross-comparisons being significantly different. For most genes the number of significantly different cross-comparisons between normal and malignant prostate tissues was similar or less than the proportion (16.7%–57.1%) of cross-comparisons between normal tissues that were significantly different, suggesting that the repositioning was not cancer specific but rather related to the variability in positioning patterns between individuals (Table 2, Figs. 2b–3). Comparing the position of genes in prostate cancer specimens to their PND also identified only a limited amount of repositioning of these genes in prostate cancer (Table 3, Figs. 2b–3). MYC, TGFB3 and HES5 repositioned in 0–33.3% prostate adenocarcinoma’s compared to their PNDs and ERBB2, FOL2, CSF1R, AKT1 and TGFB3 in 40.0–50.0% of cancer tissues compared to their PND (Table 3, Fig. 3).
The radial positions of genes in breast tissues are highly conserved between individuals (Figs. 2b–3; Meaburn et al. 2009). In contrast, we have previously identified several genes that occupy more variable positions between individuals in prostate tissue (Leshner et al. 2015). It is unclear if this is a feature of the individual loci or reflects differences in the degree of overall positional conservation between breast and prostate tissue. In keeping with their positioning patterns in breast tissue, for five of the breast GPBs there was little variability in the positioning pattern of normal prostate tissues between individuals, with 16.7%–33.3% of individual cross-comparisons amongst normal prostate tissues being significantly different (Table 2, Figs. 2b–3). Moreover, for four of these genes, none of the normal tissues were significantly different to the PND, and for TGFB3 only a single normal tissue (16.7%; 1/6 tissues) was significantly different to the PND (Table 3, Figs. 2b–3). Conversely, in contrast to their positioning profiles in breast tissue, the positions of three genes (FOL2, CSF1R and HSP90AA1) were more variable between individuals, with 38.9%–57.4% of individual cross comparisons reaching significance, and 27.3–42.9% of normal tissues significantly different to the PND (Table 2–3, Figs. 2b–3). These data suggest that differences in the extent of inter-individual variations of gene positioning patterns are tissue-rather than gene-specific.
We conclude that only two of the eleven marker genes (18.2%; MMP9 and FLI1; Suppl. Table 4) reposition in both breast and prostate cancer, demonstrating that cancer-related repositioning is not a gene-intrinsic feature but that distinct sets of genes reposition in cancer in a tissue-of-origin specific manner.
The spatial repositioning of FLI1 and MMP9 is specific to cancer
In addition to cancer, we have shown that genes can reposition in breast and prostate tissue with abnormal but non-malignant histology (Leshner et al. 2015; Meaburn et al. 2009). Therefore, to ascertain if the repositioning of the prostate GPBs markers FLI1 and MMP9 in breast cancer tissues was specific to cancer or also occurred in abnormal, but non-malignant, breast tissue, we positioned these genes in benign hyperplasia and fibroadenoma breast tissues (Table 4, Suppl. Table 2, Fig. 4). The positions of both FLI1 and MMP9 were similar in the non-malignant diseased tissues with only a single cross-comparison reaching significance for each gene (1/10; Table 4, Fig. 4). There was also only limited repositioning of FLI1 and MMP9 when the non-malignant diseased tissues were compared to normal breast tissues, with 20–26.7% of cross-comparisons to individual normal tissues and 20–40% (1/5 and 2/5, respectively) of comparisons to the PND being significant (Table 4, Fig. 4). These data suggest that FLI1 and MMP9 repositioning is specific to cancer and not a general feature of a diseased phenotype.
Table 4.
Comparison of gene positioning in benign tissues
| Gene | Tissue | % (and number) of SD cross-comparison between: | ||
|---|---|---|---|---|
| Individual benign tissues | Individual normal and benign tissues | Individual benign tissues and pooled normal | ||
| FLI1 | Breast | 10.0% (1/10) | 26.7% (8/30) | 40.0% (2/5) |
| MMP9 | Breast | 10.0% (1/10) | 20.0% (6/30) | 20.0% (1/5) |
| MMP2 | Breast | 0.0% (0/10) | 45.0% (9/20) | 40.0% (2/5) |
|
| ||||
| CSF1R | Prostate | 0.0% (0/6) | 18.2% (8/44) | 0.0% (0/4) |
SD, significantly different, based on a two-sample 1D KS test, P < 0.01.
Fig. 4.
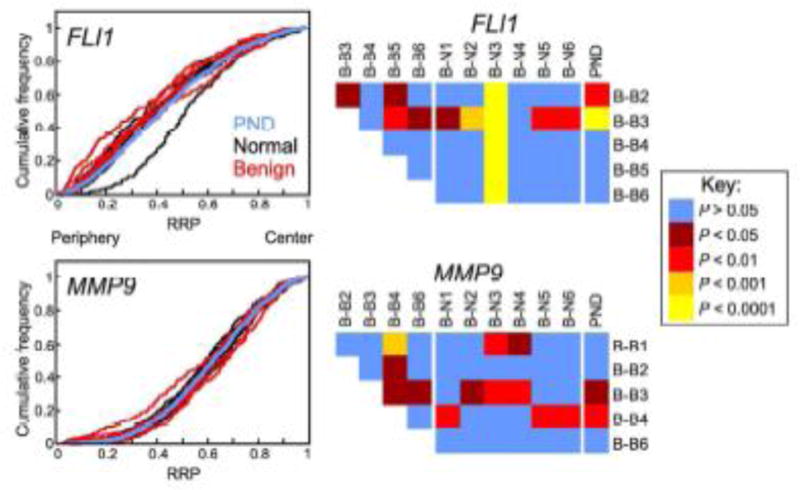
Gene positioning in non-malignant disease resembles normal tissue. Positions of indicated genes were compared between benign breast disease (hyperplasia: B-B1-B-B3; fibroadenoma: B-B4-B-B6; see Suppl. Table S2) and normal breast tissue (B-N1 – B-N6). Left-hand panel: Cumulative RRDs for the indicated genes in benign (red), normal tissues (black) and the pooled normal distribution (PND; blue). Right-hand panel: Pairwise statistical comparisons of RRDs between benign and normal tissues and amongst benign tissues, using the two-sample 1D KS test. RRP, relative radial position
Since MMP2 repositioned in hyperplasic prostate tissue (Leshner et al. 2015), we also positioned it in benign breast disease. MMP2 was positioned identically amongst all the non-malignant diseased tissues (Table 4, Suppl. Fig. S2) and showed a low level of repositioning in the non-malignant diseased breast tissues compared to normal tissue with 45.0% (9/20) of the cross-comparisons to individual normal tissues reaching significance and only two (40%) of non-malignant diseased tissues significantly different to the PND tissues (Table 4, Suppl. Fig. S2), further demonstrating the tissue-specific nature of the MMP2 repositioning event.
Normal and NAT prostate tissue often contain small areas of mild hyperplasic morphology. Therefore, as a control to ensure the heterogeneity in positioning patterns for some genes in normal prostate tissues was not a reflection of the inclusion of abnormal tissue, we positioned one of the genes with a high level of variability between individuals, CSF1R, in hyperplasic prostate tissues. CSF1R was positioned identically amongst all hyperplasic prostate tissues (Table 4, Suppl. Fig. S2). Similarly, the position of CSF1R was also similar between hyperplasic and normal tissues with all four hyperplasic prostate tissues distributions statistically similar to the PND and only 18.2% (8/44) of cross-comparisons between hyperplasic and individual normal tissues being significantly different (Table 4, Suppl. Fig. S2). This suggests that the presence of hyperplasic nuclei within a normal tissue RRD is not skewing spatial positioning patterns.
Finally, the high rate of repositioning of FLI1 and MMP9 in breast cancer tissue combined with the low level of repositioning in benign tissues make these two genes potential breast GPBs for diagnostic applications. To assess their suitability, we determined false positive and false negative rates for these two genes in breast cancer tissues (Table 5). A false positive is defined as spatial repositioning of a gene in a non-malignant tissue compared to the PND, which would incorrectly classify the tissue as cancer. Conversely, a false negative is defined as a cancer tissue where the given gene did not reposition, which would incorrectly classify the tissue as non-malignant. Both FLI1 and MMP9 had low false negative rates in breast cancer, at 0% (0/10) and 16.7% (2/12), respectively (Table 5) and had false positive rates of 27.3% (3/11) and 9.1% (1/11), respectively (Table 5). These results suggest that FLI1 and MMP9, in addition to being prostate GPBs, are also potential candidates as breast cancer biomarkers.
Table 5.
False positive and false negative rates
| Gene | Tissue | Normal tissue false positives | Benign tissue false positives | Total false positives | False negative rate |
|---|---|---|---|---|---|
| FLI1 | Breast | 16.7% (1/6) | 40.0% (2/5) | 27.3% (3/11) | 0.0% (0/10) |
| MMP9 | Breast | 0.0% (0/6) | 20.0% (1/5) | 9.1% (1/11) | 16.7% (2/12) |
| MMP2 | Breast | 25.0% (1/4) | 40.0% (2/5) | 33.3% (3/9) | 100.0% (6/6) |
|
| |||||
| AKT1 | Prostate | 0.0% (0/4) | ND | 0.0% (0/4) | 50.0% (3/6) |
| CSF1R | Prostate | 27.3% (3/11) | 0.0% (0/4) | 20.0% (3/15) | 55.6% (5/9) |
| ERBB2 | Prostate | 0.0% (0/4) | ND | 0.0% (0/4) | 60.0% (3/5) |
| FOSL2 | Prostate | 33.3% (3/9) | ND | 33.3% (3/9) | 57.1% (4/7) |
| HES5 | Prostate | 0.0% (0/6) | ND | 0.0% (0/6) | 66.7% (4/6) |
| HSP90AA1 | Prostate | 42.9% (3/7) | ND | 42.9% (3/7) | 50.0% (2/4) |
| MYC | Prostate | 0.0% (0/6) | ND | 0.0% (0/6) | 100.0% (5/5) |
| TGFB3 | Prostate | 16.7% (1/6) | ND | 16.7% (1/6) | 75.0% (3/4) |
The percentage (and number) of tissues that give a false positive or false negative result. A false positive is scored when a gene has a statistically significantly different RRD in a normal, fibroadenoma or hyperplasic tissue compared to the pooled normal (P < 0.01; KS test). A false negative is scored when a gene has a similar RRD in a cancer tissue to that of the pooled normal distribution (KS test, P > 0.01). ND, not determined.
Discussion
We report here a systematic comparison of the spatial positioning patterns of a diverse set of gene loci in both normal and malignant breast and prostate tissues. For many genes we find conservation of positioning patterns between non-malignant breast and prostate tissues. We identify only two genes that are repositioned in both breast and prostate cancer, compared to their normal counterparts. These observations demonstrate that cancer-related gene repositioning events are tissue-of-origin and gene-specific.
Consistent with previous studies in other tissues and cell types (Battulin et al. 2015; Boyle et al. 2001; Cremer et al. 2003; Foster et al. 2012; Mayer et al. 2005; Parada et al. 2004; Peric-Hupkes et al. 2010), we find considerable conservation of spatial positioning patterns between normal breast and prostate tissues, with 60% of the 15 genes analyzed in similar radial positions between the two organs. The observed differential positioning patterns of some genes between breast and prostate are unlikely to reflect global genome reorganization or repositioning of whole chromosomes, but appear to reflect changes at the level of individual genes. In support, while the position of MMP1 was dependent on tissue type, two other genes on HSA 11, FLI1 and CCND1, are similarly positioned between breast and prostate tissue. Similarly, of the three genes on HSA 14, only HSP90AA1 differs between breast and prostate tissue.
Our study extends growing evidence, from multiple tissue types including brain, thyroid, breast and prostate, that the spatial positioning of the genome is largely conserved between individuals (Borden and Manuelidis 1988; Leshner et al. 2015; Meaburn et al. 2009; Murata et al. 2007; Timme et al. 2011; Wiech et al. 2005). We find that 18 genes analyzed in multiple morphologically normal breast tissues all displayed a high level of conservation of positioning between individuals (this study and Meaburn et al. 2009). In addition, in prostate tissues 44/48 (91.7%) of genes have similar positioning patterns amongst individuals (this study and Leshner et al. 2015). However, some inter-individual variability was found, particularly in prostate. Unlike in breast (Meaburn et al. 2009), we find that radial distributions of FOSL2, CSF1R and HSP90AA1 are variable between individual prostate tissues. Similarly, the position of CCND1 was similar between breast tissues (Meaburn et al. 2009), yet variable amongst prostate tissues from multiple individuals (Leshner et al. 2015). This observation leads to the possibility that some tissues have a higher degree of intrinsic variability in spatial genome organization patterns. Due to the high prevalence of benign hyperplasia of the prostate, another possible explanation is that the variability in positioning patterns between prostates reflects a varying amount of hyperplasic nuclei included in the analysis of “normal” tissue, although our observation of similar positioning of CSF1R in hyperplasic and normal argues against this scenario. It is also possible that the extent of other benign morphologies, such as proliferative inflammatory atrophy or prostatic intraepithelial neoplasia, may affect positioning patterns between individuals.
We have previously identified eight genes that reposition in breast cancer tissues (Meaburn et al. 2009). By testing prostate GPBs in breast tissue as part of our cross-comparison, we have identified two additional genes (FLI1 and MMP9) that reposition in breast cancer, but not benign breast tissues. When we combine our current findings with our previous studies (Leshner et al. 2015; Meaburn et al. 2009), we find that 6.4% (3/47) of analyzed genes reposition in prostate cancer and 43.5% (10/23) of tested genes reposition in breast cancer. It is unclear why the proportion of repositioning genes is considerably lower in prostate cancer, although the higher inter-individual variability in prostate may be a contributing factor. Future studies employing high-throughput imaging to map a larger number of genes should determine if this difference is due to the limited number of tested genes or reflects underlying biological properties or both. The molecular properties that determine whether a gene repositions between normal and cancer tissues are currently unknown. Analysis of repositioned genes in breast and prostate cancer has demonstrated that cancer-related repositioning does not correlate with their gene expression status, gene copy number or local gene density (Leshner et al. 2015; Meaburn et al. 2009; Meaburn and Misteli 2008).
We find that a particular gene can behave differently in cancers from different tissues. Of the eleven genes previously identified to reposition in either breast (Meaburn et al. 2009) or prostate cancer (Leshner et al. 2015), only two (FLI1 and MMP9), spatially reposition in both breast and prostate cancer. In keeping with our finding, BCL2 has previously been identified to reposition in some cervical squamous carcinomas (Wiech et al. 2009), but not breast (Meaburn et al. 2009) or prostate (Leshner et al. 2015) cancer. This suggests that distinct groups of genes reposition in cancers from different tissues. While breast and prostate cancer share biological features such as their origin from epithelial cells and dependence on sex steroid hormones (Risbridger et al. 2010), it will be interesting to determine in future studies how general the repositioning of FLI1 and MMP9 is in other cancers.
The fact that different genes spatially reposition in different cancer types may have implications for diagnostics as it suggests a correlation between non-random repositioning of the genome during carcinogenesis with the specific biology of the cancer. This leads to the possibility that differential positioning patterns for specific genes may occur even within cancers originating from the same tissue, including amongst sub-types of tumors. Identification of sub-type specific positioning biomarkers may be valuable in cancer prognostics, for example in distinguishing indolent from aggressive cancers.
Supplementary Material
Acknowledgments
We thank Prabhakar Gudla and Stephen Lockett for assistance with image analysis; Delft University, Netherlands for providing the DIPImage and PRTools toolboxes; and Tatiana Karpova for microscopy support. Fluorescence imaging was performed at the National Cancer Institute Fluorescence Imaging Facility. We also thank Ms. Kathy Doan for help in identifying and obtaining prostate tissue used for this study.
Funding: This work was supported by a Department of Defense Idea Awards (W81XWH-12-1-0224 and W81XWH-12-1-0295), by NCI, National Institutes of Health (NIH) under award Pacific Northwest Prostate Cancer SPORE grant (P50CA097186), and in part by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Abbreviations
- BAC
bacterial artificial chromosome
- EDT
Euclidean distance transform
- FFPE
formalin-fixed, paraffin embedded
- GPBs
gene positioning biomarkers
- HSA
human chromosome
- KS test
Kolmogorov-Smirnov test
- MMU
mouse chromosome
- NAT
Normal adjacent to tumor
- PND
pooled normal distribution
- RRD
relative radial distribution
- TMA
tissue microarray
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent: Informed consent was obtained from all individual participants included in the study.
References
- Battulin N, et al. Comparison of the 3D organization of sperm and fibroblast genomes using the Hi-C approach. Genome biology. 2015;16:77. doi: 10.1186/s13059-015-0642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden J, Manuelidis L. Movement of the X chromosome in epilepsy. Science. 1988;242:1687–1691. doi: 10.1126/science.3201257. [DOI] [PubMed] [Google Scholar]
- Boutanaev AM, Mikhaylova LM, Nurminsky DI. The pattern of chromosome folding in interphase is outlined by the linear gene density profile. Mol Cell Biol. 2005;25:8379–8386. doi: 10.1128/MCB.25.18.8379-8386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Cremer M, et al. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. The Journal of cell biology. 2003;162:809–820. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, de Castro IJ, Lavitas L, Chotalia M, Pombo A. Gene positioning Cold Spring Harbor perspectives in biology. 2010;2:a000588. doi: 10.1101/cshperspect.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster HA, Griffin DK, Bridger JM. Interphase chromosome positioning in in vitro porcine cells and ex vivo porcine tissues. BMC cell biology. 2012;13:30. doi: 10.1186/1471-2121-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hiratani I, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kurz A, et al. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshner M, Devine M, Roloff GW, True LD, Misteli T, Meaburn KJ. Locus-specific gene repositioning in prostate cancer. Molecular biology of the cell. 2015 doi: 10.1091/mbc.E15-05-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. The Journal of cell biology. 2002;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, Dietzel S. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC cell biology. 2005;6:44. doi: 10.1186/1471-2121-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ. Fluorescence in situ hybridization on 3D cultures of tumor cells. Methods in molecular biology. 2010;659:323–336. doi: 10.1007/978-1-60761-789-1_25. [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Burman B, Misteli T. Spatial genome organization and disease. In: Dellaire G, Basett-Jones D, editors. The Functional Nucleus. Springer DE; 2016. In press. [Google Scholar]
- Meaburn KJ, et al. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging cell. 2007;6:139–153. doi: 10.1111/j.1474-9726.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Gudla PR, Khan S, Lockett SJ, Misteli T. Disease-specific gene repositioning in breast cancer. The Journal of cell biology. 2009;187:801–812. doi: 10.1083/jcb.200909127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Locus-specific and activity-independent gene repositioning during early tumorigenesis. The Journal of cell biology. 2008;180:39–50. doi: 10.1083/jcb.200708204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Newbold RF, Bridger JM. Positioning of human chromosomes in murine cell hybrids according to synteny. Chromosoma. 2008;117:579–591. doi: 10.1007/s00412-008-0175-3. [DOI] [PubMed] [Google Scholar]
- Mehta IS, Eskiw CH, Arican HD, Kill IR, Bridger JM. Farnesyltransferase inhibitor treatment restores chromosome territory positions and active chromosome dynamics in Hutchinson-Gilford progeria syndrome cells. Genome biology. 2011;12:R74. doi: 10.1186/gb-2011-12-8-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewborn SK, et al. Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLoS ONE. 2010;5:e14342. doi: 10.1371/journal.pone.0014342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikelsaar R, Paves H, Org K, Mikelsaar AV. Chromosome variant 1qh- and its influence on the 3D organization of chromosome 1 heterochromatin in interphase nucleus of patients with endometriosis. Journal of genetics. 2014;93:219–223. doi: 10.1007/s12041-014-0340-9. [DOI] [PubMed] [Google Scholar]
- Murata S, et al. Conservation and alteration of chromosome territory arrangements in thyroid carcinoma cell nuclei. Thyroid: official journal of the American Thyroid Association. 2007;17:489–496. doi: 10.1089/thy.2006.0328. [DOI] [PubMed] [Google Scholar]
- Parada L, McQueen P, Misteli T. Tissue-specific spatial organization of genomes. Genome biology. 2004;7:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz N, et al. Expression of the DYRK1A gene correlates with its 3D positioning in the interphase nucleus of Down syndrome cells. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2015 doi: 10.1007/s10577-015-9467-7. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbridger GP, Davis ID, Birrell SN, Tilley WD. Breast and prostate cancer: more similar than different. Nat Rev Cancer. 2010;10:205–212. doi: 10.1038/nrc2795. [DOI] [PubMed] [Google Scholar]
- Scheuermann MO, Tajbakhsh J, Kurz A, Saracoglu K, Eils R, Lichter P. Topology of genes and nontranscribed sequences in human interphase nuclei. Exp Cell Res. 2004;301:266–279. doi: 10.1016/j.yexcr.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Shopland LS, et al. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. The Journal of cell biology. 2006;174:27–38. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme S, et al. Nuclear position and shape deformation of chromosome 8 territories in pancreatic ductal adenocarcinoma. Analytical cellular pathology. 2011;34:21–33. doi: 10.3233/ACP-2011-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- Volpi EV, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Wiech T, et al. Spatial allelic imbalance of BCL2 genes and chromosome 18 territories in nonneoplastic and neoplastic cervical squamous epithelium. Eur Biophys J. 2009;38:793–806. doi: 10.1007/s00249-009-0474-5. [DOI] [PubMed] [Google Scholar]
- Wiech T, et al. Human archival tissues provide a valuable source for the analysis of spatial genome organization. Histochem Cell Biol. 2005;123:229–238. doi: 10.1007/s00418-005-0768-3. [DOI] [PubMed] [Google Scholar]
- Williams RR, Broad S, Sheer D, Ragoussis J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp Cell Res. 2002;272:163–175. doi: 10.1006/excr.2001.5400. [DOI] [PubMed] [Google Scholar]
- Zeitz MJ, Ay F, Heidmann JD, Lerner PL, Noble WS, Steelman BN, Hoffman AR. Genomic interaction profiles in breast cancer reveal altered chromatin architecture. PLoS ONE. 2013;8:e73974. doi: 10.1371/journal.pone.0073974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, et al. Transcription-dependent spatial arrangement of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;I166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


