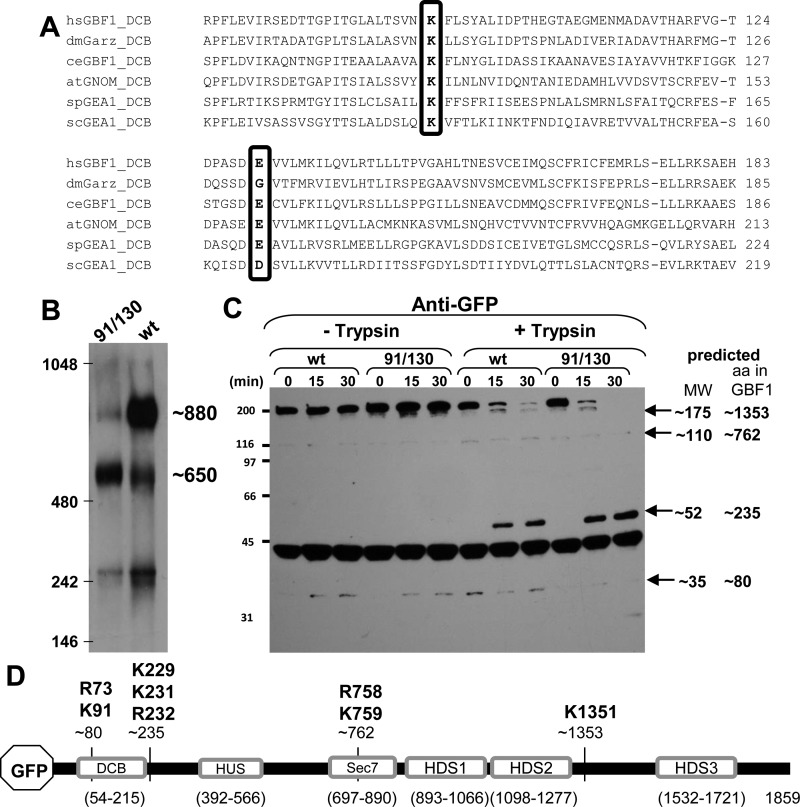
Fig. 1.
Oligomerization of GBF1 requires K91 and E130. A: alignment of the dimerization and cyclophilin binding (DCB) domains of GBF1 orthologs from various organisms show conservation of K91 and E130 (boxed). Numbering according to the human GBF1 sequence. B: lysates prepared from HeLa cells expressing either green fluorescent protein (GFP)-tagged GBF1 (GFP-GBF1) or GFP-91/130 were separated on blue native gels, transferred to PVDF membranes, and immunoblotted with anti-GFP. GFP-GBF1 (wt) predominantly migrates at ∼880 kDa, while GFP-91/130 predominantly migrates at ∼650 kDa. C: lysates prepared from HeLa cells expressing either GFP-GBF1 or GFP-91/130 were incubated without or with 2.5 μg/ml trypsin for indicated times, separated by SDS-PAGE, and processed for immunoblotting with anti-GFP. Time-dependent proteolysis of both constructs is detected in the presence of trypsin. The predicted molecular weights of the fragments are indicated. D: schematic of GFP-GBF1 showing the domains and the approximate cleavage sites generating the fragments detected in C. The amino acid residues indicated above the fragments represent the bioinformatically predicted putative cleavage sites for trypsin. HUS, homology upstream of Sec7; HDS, homology downstream of Sec7.