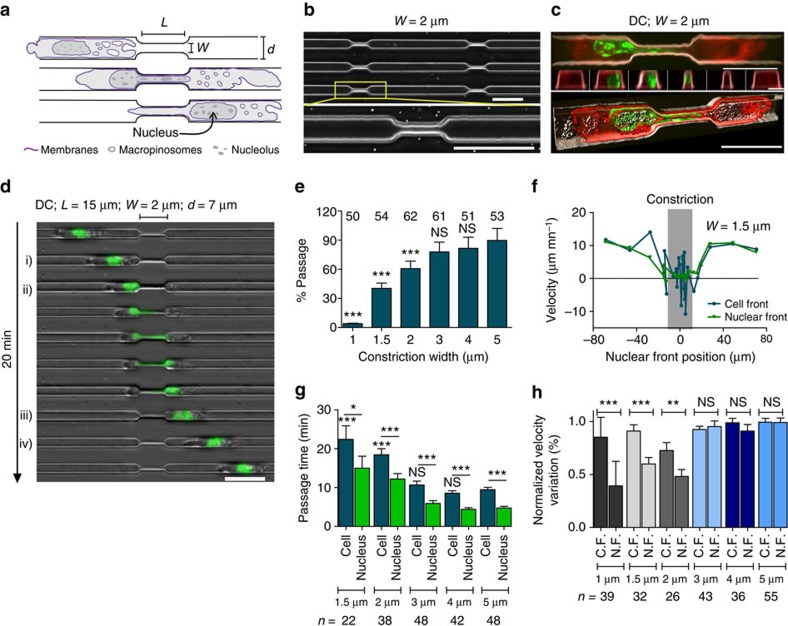
Figure 1. The nucleus imposes a physical limitation to DC migration through micrometric pores.
(a) Schematic representation of the experimental setup. (b) Top: representative image of a field of channels with constrictions. Bottom: zoom on a single constriction. Scale bar, 30 μm. (c) Top: representative image of an Iaβ-GFP (red)-expressing DC stained with Hoechst (DNA, green), passing through a 2-μm constriction coated with pLL-PEG-Rhodamin (grey levels). Scale bar, 15 μm. Middle: views of the channel cross-section before, along and after the constriction. Scale bar, 4 μm. Bottom: perspective view (45°) of a 3D iso-surface reconstruction. Scale bar, 15 μm. (d) Representative sequential images of a DC stained with Hoechst (green) migrating through a constriction. (i) Cell entry, (ii) nuclear entry, (iii) nuclear exit and (iv) cell exit. Scale bar, 30 μm. Time in minutes: seconds. (e) Percentage of passage through 20-μm-long constrictions. Numbers above bars represent the number of cells scored. (f) Representative cell (blue) and nuclear (green) front instantaneous velocity as a function of the nuclear front position in 1.5 μm constrictions.(g) Cell (blue) and nuclear (green) passage time in 20-μm-long constrictions. (e,g) Unless when indicated by a spanner, statistical test was against the value for 5-μm-wide constrictions. (h) Cell and nuclear front velocities inside constrictions, normalized by the cell centre of mass velocity before the constriction. C.F., cell front; N.F., nuclear front. Error bars represent the s.e.m. (g,h) n represents the number of cells. ***P-value<0.0001, **P-value<0.001 and *P-value<0.01; NS,=nonsignificant. Statistical test: Fisher's test for e, Mann–Whitney test for g and h. Error bars are s.e.m. (a,d) L and W for constriction length and width, respectively; d for channel width; N≥2.