Abstract
Phylogeny reflects genetic and phenotypic traits in Bacteria and Archaea. The phylogenetic conservatism of microbial traits has prompted the application of phylogeny-based algorithms to predict unknown trait values of extant taxa based on the traits of their evolutionary relatives to estimate, for instance, rRNA gene copy numbers, gene contents or tolerance to abiotic conditions. Unlike the ‘macrobial' world, microbial ecologists face scenarios potentially compromising the accuracy of trait reconstruction methods, as, for example, extremely large phylogenies and limited information on the traits of interest. We review 990 bacterial and archaeal traits from the literature and support that phylogenetic trait conservatism is widespread through the tree of life, while revealing that it is generally weak for ecologically relevant phenotypic traits and high for genetically complex traits. We then perform a simulation exercise to assess the accuracy of phylogeny-based trait predictions in common scenarios faced by microbial ecologists. Our simulations show that ca. 60% of the variation in phylogeny-based trait predictions depends on the magnitude of the trait conservatism, the number of species in the tree, the proportion of species with unknown trait values and the mean distance in the tree to the nearest neighbour with a known trait value. Results are similar for both binary and continuous traits. We discuss these results under the light of the reviewed traits and provide recommendations for the use of phylogeny-based trait predictions for microbial ecologists.
Trait-based approaches in community ecology studies are becoming increasingly appealing for microbial ecologists partly because metagenomic sequencing allows surveying molecular functions (Green et al., 2008; Lauro et al., 2009; Burke et al., 2011; Raes et al., 2011; Brown et al., 2014; Fierer et al., 2014). Although genetic data can provide precise information on cellular processes or metabolic pathways, they are generally blind to other ecologically relevant phenotypic traits such as the tolerance to certain abiotic conditions or the specific growth rate (but see Vieira-Silva and Rocha, 2010). Unlike ‘macrobial' ecologists, who can directly observe phenotypic characters of plants and animals, microbial ecologists usually face situations where most of the phenotypes of their study organisms are unknown. This difficulty relies on the fact that gathering phenotypic (physiological, morphological, biochemical) data requires culturing microbial species. The unbalanced growth of genotypic vs phenotypic information is currently challenging microbial ecologists to work with phylogenetic trees of increasing size (hundreds to thousands of species) in which the percentage of species with unknown traits becomes larger and larger.
Recent evidence indicate that phylogeny reflects molecular functions and phenotypes in Bacteria and Archaea (Langille et al., 2013; Martiny et al., 2013). This is due to the phylogenetic conservatism of microbial traits (Martiny et al., 2013), which likely arises from microbial evolution mostly proceeding by vertical gene inheritance rather than horizontal gene transfer (Kurland et al., 2003, see Fraser et al., 2007 for theoretical models on the role of horizontal gene transfer in bacterial speciation). At present, the massive sequencing of microbes in the environment is providing a huge amount of genetic information that is extremely useful to reconstruct the phylogenetic relationships among microbial lineages. This fact has triggered the interest of microbial ecologists to apply the methods developed to predict unobserved trait values of extant taxa based on the traits observed in their evolutionary relatives (Kembel et al., 2012; Langille et al., 2013; Angly et al., 2014, see review in Zaneveld and Thurber, 2014). All these methods are based on the existence of a significant phylogenetic signal or, in other words, in the fact that close relatives have more similar traits than expected by chance. Phylogeny-based trait prediction procedures (PTP hereafter) in microbes have been mainly performed under the phylogenetic generalized least squares framework (Martins and Hansen, 1997; Garland and Ives, 2000). Specifically, the trait value (for continuous traits) or state (for binary traits) of the focal species have been reconstructed through ancestral state reconstructions after rerooting the phylogeny at the most recent common ancestor of the taxon with unobserved trait and the rest of the tree (Kembel et al., 2012). The accuracy of PTP methods has been typically assessed under ‘macrobial' scenarios containing phylogenies of moderate size, with low-to-medium proportion of species with unknown traits and significant phylogenetic signals. For example, Fagan et al. (2013) predicted population growth rates of mammals in phylogenies of 42–65 species containing 54–64% of unknowns and a significant phylogenetic signal (Blomberg et al., 2003; Blomberg's K) ranging from 0.68 to 1.42. However, the current microbial scenarios derived from high-throughput sequencing projects face large-sized phylogenies (hundreds to thousands tips) with a high number of species with unknown traits and varying phylogenetic signals jeopardizing the applicability of PTP methods (Zaneveld and Thurber, 2014).
The extent to which phylogeny reflects phenotype is strongly dependent on the degree of conservatism with which the focal trait has evolved. For instance, complex traits that involve many genes (for example, photosynthesis or methanogenesis) show higher conservatism than simpler traits, such as the consumption of a specific carbon source (Martiny et al., 2013). Furthermore, certain traits such as those related to genes encoding antibiotic or metal resistance are particularly prone to be horizontally transferred (Bruins et al., 2000), a process that can blur their phylogenetic signal. Therefore, if phylogenetic relatedness is to be used to infer the phenotype, the phylogenetic conservatism of the target trait needs to be quantified in every case.
Altogether, the abovementioned observations indicate that the possibility to estimate phenotypes from phylogenies depends on the amount of phylogenetic and phenotypic information available to predict the unobserved trait values. Here we provide a simulation exercise to test the accuracy of the most widely used PTP method in microbial ecology to predict continuous trait values and binary trait states of extant taxa with different amount of phenotypic and phylogenetic information. We simulated several situations faced by microbial ecologists, including phylogenies of different sizes in which a small (P=0.3), medium (P=0.6) or large (P=0.9) proportion of species have unknown trait values. The correlations between the actual and the predicted trait values were obtained for characters evolved under different degree of conservatism. Finally, we put these values in the context of the phylogenetic signals described in the literature for different continuous and binary microbial traits and provide some recommendations for future analyses aimed to predict microbial traits with the help of the phylogenetic information.
Materials and methods
Simulations of phylogeny-based trait predictions
The accuracy of trait predictions under different scenarios were studied following four sequential steps: (i) simulating trait evolution in a phylogenetic tree, (ii) removing trait values from a number of species in the tree, (iii) reconstructing the trait values in the species previously removed, and (iv) comparing the actual with the predicted trait values.
Five hundred trees for each combination of trait type (continuous and binary) × number of species (100 and 1000) × proportion of unknown unobserved traits of species in the phylogeny (30, 60 and 100%) were generated by simulating stochastic pure birth trees with the pbtree command in the phytools package for R (Revell, 2012). Pure-birth has been shown to be a convenient model describing the evolution of bacterial lineages (Lorén et al., 2014).
The evolution of continuous traits with different strength of phylogenetic signal was simulated in the phylogenetic trees generated above. A phylogenetic signal equalling the evolution under a Brownian Motion (BM) expectation was obtained with the fastBM function in the phytools package. Phylogenetic signals departing from BM were obtained by adding increased amounts of random noise to the trait vector generated by fastBM. Random noise was generated through a normal distribution with mean=0 and different values of s.d. Large s.d. values increase random noise and reduce the phylogenetic signal of the trait. To obtain phylogenetic signals higher than the BM expectation, we used the high-signal-trait R code developed by Steve Kembel (https://gist.github.com/skembel/8523702; accessed 14 April 2015). This code generates a highly conserved trait by scaling phylogeny branch lengths with a delta time-dependent model of trait evolution (Pagel, 1999). Slow trait evolution, and therefore high phylogenetic signal, is obtained when delta values are <1. Phylogenetic signals of continuous traits were calculated with Blomberg's K statistic (Blomberg et al., 2003) in the picante package. This test compares the variance of the phylogenetically independent contrast of the study trait against those obtained with data randomly reshuffled in the phylogeny. It quantifies the phylogenetic signal in the interval (0, ∞) indicating whether the evolution of a trait (a) does not show a significant signal (K=0); (b) is more conserved than expected by chance (K>0); (c) is less conserved than expected under BM (0<K<1), (d) is as conserved as expected under BM (K=1) or (e) is more conserved than expected under BM (K>1).
The evolution of binary traits along the phylogenetic trees was simulated through a Markovian model where the transition probabilities between the states of the trait are calculated for each branch. This procedure was run with the help of the rTraitDisc function in the ape package for R (Paradis et al., 2004). Different phylogenetic signal strengths were obtained by altering the transition probabilities between states. The high-signal-trait R code was also used to generate binary traits with high phylogenetic signals. Phylogenetic signal of binary traits was calculated with the phylo.D algorithm in the caper package (Orme et al., 2013). This metrics compares the observed sister-clade differences in the study trait against those expected for a random phylogenetic pattern. It was developed by Fritz and Purvis (2010) to calculate the strength of the phylogenetic signal in binary traits and ranges within the interval (−∞, ∞), with low values indicating trait conservatism. However, for comparative purposes with Blomberg's K statistic, we transformed the D value into −D+1 to indicate whether the evolution of a trait (a) does not show a significant signal (−D+1=0); (b) is more conserved than expected by chance (−D+1>0); (c) is less conserved than expected under BM (0<−D+1<1), (d) is as conserved as expected under BM (−D+1=1) or (e) is more conserved than expected under BM (−D+1>1).
Once we had the traits evolved onto the phylogenetic trees, we randomly removed the trait states of a different proportion of species (30, 60 and 90%) in the trees. To account for the closeness of these removed species to the nearest relative with known traits, we calculated their mean nearest neighbour phylogenetic distance (MNND). We subsequently reconstructed these missing traits with the help of the functions phyEstimate for continuous traits and phyEstimateDisc for binary traits in the picante package for R (Kembel et al., 2010). These functions use phylogenetic ancestral state estimation to infer trait values for novel taxa on a phylogenetic tree by rerooting the tree on the parent edge for the node to be predicted (Kembel et al., 2012). Finally, the accuracy of trait reconstructions was obtained by correlating the predicted values obtained with these algorithms with the actual values of the traits. Pearson correlations were used for continuous traits and Spearman rank correlations for binary traits.
To mimic a more realistic situation where the availability of microbial data are clustered around particular groups of interest (that is, human pathogens or organisms of biotechnological interest), we also simulated several scenarios where species with unknown traits were not randomly distributed in the phylogeny but clustered within certain clades. We performed the same procedure as described above but pruning the desired percentage of tips (30, 60 or 90%) within particular clades instead of randomly pruning across clades. These clades were detected with the help of the getCladesofSize command in the phytools package for R (Revell, 2012). This function gets all subtrees that cannot be further subdivided into two reciprocally monophyletic subtrees of size higher than a given clade size. We set to five the clade size, producing extremely clustered pruning. PTP was subsequently performed for traits evolved within the range of phylogenetic signals for bacterial and archaeal traits observed in the literature, as explained below.
To evaluate the factors affecting the accuracy of trait prediction in our simulations, we fitted a linear model with the correlation between predicted and actual trait values as the dependent variable and phylogenetic signal, number of species in the phylogeny, the proportion of unknown species as independent variables and MNND. The phylogenetic signal (both K and −D+1) were log-transformed to account for the non-linear relationship with the dependent variable.
Phylogenetic signals of Bacteria and Archaea
The search of studies quantifying phylogenetic signals of microbial traits was performed by using a combination of the keywords ‘phylogenetic signal' and ‘bacteria' and/or ‘archaea' in Web of Science and Google Scholar until February 2015. Very few studies were found and then we enlarged our database by calculating phylogenetic signals when trait information was provided at the strain or species level. In cases where the tree reconstructing the phylogenetic relationships among species was not given, we assumed the topology of the Silva tree (Release 119, Quast et al., 2013). In brief, we exported the guide tree containing >125 000 sequences of cultured organisms with the ARB software package (Ludwig et al., 2004). We randomly removed duplicate names (that is, sequences belonging to the same organism) from the guide tree and further pruned it to obtain smaller trees containing the set of organisms for which trait information was available in each case using the ape package for R (Paradis et al., 2004). These phylogenetic trees and the corresponding trait data are provided in Supplementary Information S1 as an RData object.
We also computed the phylogenetic depth at which binary traits were conserved with the help of the consenTrait index (Martiny et al., 2013). This index determines the mean depth of clades containing >90% of species sharing a trait.
Results
Simulations of phylogeny-based trait predictions
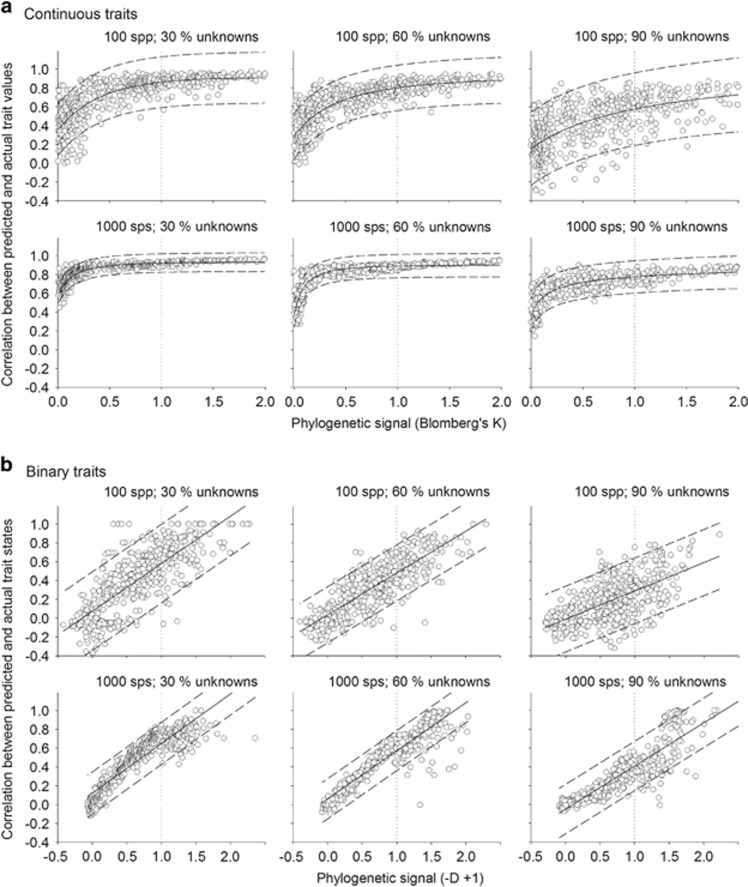
When continuous traits were evolved in the simulated phylogenetic trees, the correlation between the predicted and the actual trait values increased in a non-linear manner with the phylogenetic signal of the trait, measured as Blomberg's K (Figure 1a). The accuracy of trait prediction increased very fast from low (K~0.1) to medium (K~0.5) phylogenetic signals and tended to stabilize around the phylogenetic signal equalling the BM expectation (K=1). For binary traits, the correlation between the predicted and the actual trait states increased linearly with the phylogenetic signal of the trait, measured as −D+1, with the exception of the best scenario (1000 species and only 30% of unknowns) where a non-linear trend appeared (Figure 1b).
Figure 1.
Accuracy of the PTP method for (a) continuous and (b) binary traits evolved in simulated phylogenetic trees with varying numbers of species in the tree (100, 1000) and proportions of unknown species (30, 60, 90%). The magnitude of the phylogenetic signal increases towards the positive pole of both statistics (K and −D+1). Solid and dashed lines represent the regression slope and prediction intervals. Dotted lines indicate phylogenetic signals equalling a BM expectation.
The accuracy of the predictions decreased with the proportion of species with unknown trait values. For example, in small phylogenies (N=100), the expected correlation between the actual and the reconstructed continuous trait values under BM (that is, K=1; vertical dotted lines in Figure 1a, upper panel), was 0.88, 0.80 and 0.57 for the scenarios where 30, 60 and 90% of the species had unknown trait values, respectively. Similarly, for binary traits the correlation between the predicted and the actual trait states under BM (that is, −D+1=1; vertical dotted lines in Figure 1b, upper panel) was 0.58, 0.48 and 0.29 for the scenarios with 30, 60 and 90% of unknowns, respectively. The uncertainty associated with this relationship increased with the proportion of species whose traits were unknown, as shown by the large scattering and the wider prediction intervals in the plots with high proportion of species with unknown traits (Figure 1). The same trends were observed with large phylogenies (N=1000), although the predictions under BM were more accurate as shown by the higher correlations between actual and reconstructed states (continuous traits: 0.92, 0.89 and 0.77 (Figure 1a lower panel); binary traits: 0.65, 0.57 and 0.41 (Figure 1b lower panel) for the scenarios where 30, 60 and 90% of the species had unknown traits, respectively) and the narrower prediction intervals.
Taking together all the factors, the accuracy of trait prediction significantly increased with the phylogenetic signal and the number of species in the phylogeny and decreased with the percentage of unknown species and MNND (Table 1). The negative sign of MNND in the model indicates that the lower the distance the higher the accuracy of trait prediction. Altogether, these four variables explained 63% and 59% of the variation in the accuracy of the trait prediction in continuous and binary traits, respectively (Table 1). Phylogenetic signals and number of species yielded the highest contributions to the total variation in the model while the proportion of unknowns and MNND explained a very low percentage of variance. However, MNND may become relevant in other scenarios where species with unknown traits are not randomly distributed but extremely clumped in the phylogeny, as will be shown at the end of the following section.
Table 1. Linear model fits to explain the correlation between predicted and actual continuous (top panel) and binary (bottom panel) trait values as a function of different variables in the simulated phylogenetic trees.
| Estimate±s.e. | t | Variance explained | |
|---|---|---|---|
| Continuous traits | |||
| (Intercept) | 9.0E-01±7.3E-03 | 122.9* | |
| log(phylogenetic signal) | 1.1E-01±1.9E-03 | 57.4* | 40.6% |
| Proportion of unknown species | −1.2E-01±2.3E-02 | −5.1* | 0.32% |
| Number of species | 2.2E-04±5.4E-06 | 40.4* | 20.2% |
| MNND | −7.3E-02±5.9E-03 | −12.3* | 1.9% |
| Binary traits | |||
| (Intercept) | 1.0e+00±1.2E-02 | 79.1* | |
| log(phylogenetic signal) | 7.1e-01±1.1E-02 | 63.8* | 55.5% |
| Proportion of unknown species | −1.5e-01±2.6E-02 | −5.9* | 0.5% |
| Number of species | 9.5e-05±7.6E-06 | 12.5* | 2.1% |
| MNND | −6.4e-02±7.3E-03 | −9.08* | 1.1% |
Abbreviation: MNND, mean distance of the species with unknown traits to their nearest relatives. *P<0.001.
Phylogenetic signals of Bacteria and Archaea
Our data set contained phylogenetic signals for 990 microbial traits compiled from the literature, most of which (91%) were binary traits (Supplementary Information S2 and S3). A total 90% of all binary traits described molecular functions, specifically the presence or absence of a gene or a set of genes involved in biochemical pathways. The remaining 10% described phenotypic traits mostly related to tolerance to abiotic conditions, ecological interactions and consumption of specific organic substances (Supplementary Information S2). A total 61% of the continuous traits were related to genomic characteristics, such as GC content, genome size or copy numbers of specific genes, while 39% were phenotypic traits related to cellular features, response to the environment and growth rate (Supplementary Information S3). Phylogenetic signals in the literature were calculated at different taxonomic levels. Most study cases (86%) targeted simultaneously two domains (Bacteria and Archaea), while the rest were computed for the bacterial domain (9%), a single phylum (4%) or a functional group (1%).
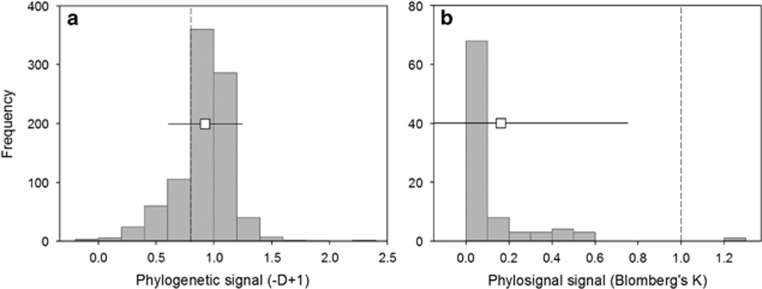
Binary microbial traits showed a mean phylogenetic signal of −D+1=0.93±0.01 (Figure 2a). A total 98% of these signals (878 out of 899) were significant indicating trait conservatism. Most of the traits were conserved at very shallow clade depths (Supplementary Information S2). The mean phylogenetic signal of continuous microbial traits was K=0.16±0.59 (Figure 2b). Seventy-four percent (67 out of 91) of these traits showed significant phylogenetic signals. Taking the average, phylogenetic signal of continuous traits compiled from the literature as a reference (K=0.16) and according to our simulations PTP would perform well in the best scenario of 1000 species and 30% unknowns (r ranging from 0.70 to 0.87) but not in the scenario with limited information (100 species and 90% unknowns; r from −0.12 to 0.66; Figure 1a). Similarly, for binary traits, whose mean phylogenetic signal shown by microbial traits was −D+1=0.93, predictions would be better for scenarios with more information (that is, 1000 species and 30% unknowns; r from 0.38 to 0.84) than for scenarios with limited information (that is, 100 species and 90% unknowns; r from −0.08 to 0.62; Figure 1b). PTP would be recommendable for continuous traits with phylogenetic signals K⩾0.4 (for example, for K=0.4, r ranged from 0.78 to 0.97 for 1000 species and 30% unknowns; Figure 1a). Reaching similar predictions with binary traits would require phylogenetic signals of –D+1⩾1.14 (for example, for –D+1=1.14, r ranged from 0.52 to 0.95 for 1000 species and 30% unknowns; Figure 1b). In our data set, these acceptable conditions would apply to 9.9% and 8.6% of the continuous and binary traits, respectively (Supplementary Information S2 and S3). Fortunately, the prediction accuracy of PTP even for a high percentage of unknowns will increase as larger phylogenies become available as simulations in trees containing up to 10 000 species show (Supplementary Information S4).
Figure 2.
Phylogenetic signals of 990 microbial (a) binary and (b) continuous traits compiled from the literature. The magnitude of the phylogenetic signal increases towards the positive pole of both statistics (−D+1 and K). Boxplots indicate average and s.d. values. Dashed lines indicate phylogenetic signals equalling a BM expectation. The signals of three binary traits with –D+1⩾3 and one continuous trait with K>5 were not included for clarity.
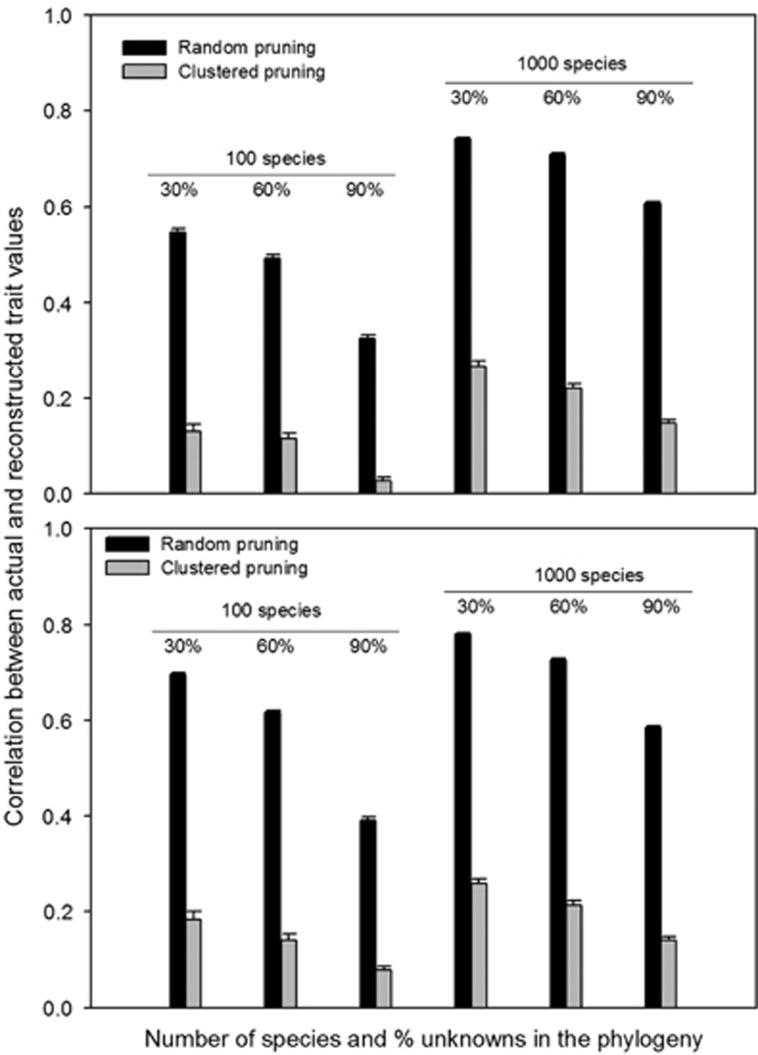
In the extreme situation where the availability of trait data were clustered in particular clades of the phylogenetic tree, the accuracy of PTP was drastically reduced (Figure 3). By pruning whole clades, the average distance of the species with unknown traits to their nearest relatives with known traits was greater by fourfold in our simulated trees.
Figure 3.
The accuracy of PTP diminishes drastically when the trait values of the known species are phylogenetically clustered in both continuous (top panel) and binary (bottom panel) traits evolved with the most frequent phylogenetic signals of bacterial and archaeal traits. The trend was similar across all the simulated scenarios.
Discussion
Our literature review confirms that the presence of a phylogenetic signal in microbial traits is widespread across the bacterial and archaeal tree of life (Martiny et al., 2013). This observation implies that the phylogenetic conservatism of traits is a universal phenomenon that has been previously reported for eukaryotes (Freckelton et al., 2002; Blomberg et al., 2003). Comparatively, however, prokaryotes show continuous traits with weak phylogenetic signals. As an illustration, the average signal that we found in the literature for continuous bacterial and archaeal traits (K~0.11) was remarkably lower than the average reported for plants and animals (K~0.77) by Blomberg et al. (2003). Differences might be partly methodological and specifically related to the taxonomic breadth at which phylogenetic signals are computed. Although in eukaryotes calculations are performed at narrow taxonomic levels (from genus to subphylum, Blomberg et al., 2003), the vast majority of studies that we compiled considered a whole domain (Bacteria) or even two domains (Bacteria and Archaea). Working at such a broad taxonomic resolution can decrease the magnitude of the phylogenetic signal as microbial traits tend to be conserved at shallower clade depths (Martiny et al., 2013; see Supplementary Information S5 for examples with simulated and empirical data). The weaker phylogenetic signals detected in prokaryotes compared with eukaryotes can also arise from the horizontal transfer of genes, which is a remarkable natural source of variation (Dagan et al., 2008). This might weaken the signal by partly shuffling the traits in the phylogeny but does not blur it as the fixation of horizontally transferred genes is not substantial between phylogenetically distant microbes (Choi and Kim, 2007). Other numerous evolutionary processes show complex relationships with the phylogenetic signal of traits as has been thoroughly discussed (Revell et al., 2008). Here we rather focus on the use of phylogenetic signal as an evolutionary pattern that can allow predicting microbial traits from phylogenies.
An interesting outcome of our phylogenetic signal database is that the most conserved continuous traits (for example, optimal pH and temperature for growth, soil moisture niche breadth or lag time prior to exponential growth) are abiotic stress tolerance and competition-related traits that can underlie microbial community assembly (Goberna et al., 2014). These are probably complex traits controlled by multiple genes and their interactions. Genetically complex traits in microbes are evolutionarily conserved at very deep phylogenetic depths, resulting in stronger phylogenetic signals (Martiny et al., 2013) and consequently allowing for more accurate predictions of traits as our simulations demonstrate. Supporting this view that genetically complex traits are evolutionarily conserved, we found that most of binary traits reflecting the presence or absence of metabolic pathways are strongly conserved.
Caution is needed to reconstruct traits for most of the microbial traits where a low phylogenetic signal has been detected. In these cases, the question arises whether it is convenient to use phylogeny-based trait predictions. Our simulation results suggest that the answer depends on three main factors other than the magnitude of the phylogenetic signal, namely, the number of species in the phylogeny, the proportion of species with unknown trait values and the distance of these species to their nearest neighbors with known traits. In brief, our ability to predict traits with phylogenies increased as traits were phylogenetically more conserved, trees were more populated and there was a higher proportion of species with known trait values. The efforts of microbial ecologists to improve the ability to predict traits should be then addressed to enlarge the phylogenetic tree of phenotyped Bacteria and Archaea. For the observed phylogenetic signals of the studied traits, phylogenies including >2000 tips allow to reconstruct unknown traits very accurately. These efforts to increase the phylogenetic knowledge of traits with moderate signals, such as soil moisture niche breath, lag time prior to exponential growth, optimal pH for growth or salt tolerance, might push the correlations between the observed and the actual trait values to r=0.65–0.90. It should be also noted that this would be the case if our phylogenetic knowledge was randomly or evenly distributed across the whole phylogenetic tree. However, much of our current knowledge is clustered around organisms of medical and biotechnological interest. As our simulations show, under an extremely clustered situation the accuracy of trait prediction is drastically reduced. Although the current situation is probably not as extreme as depicted in our clustered scenario, it is of special importance to extend our phenotypic knowledge across all the clades of the phylogenetic tree guided by a phylogenetic-diversity-driven genome sequencing approach (Wu et al., 2009; Rinke et al., 2013; Shih et al., 2013).
Possible biases affecting PTP, as those emerging from taxonomic sampling bias, intraspecific variability or trait-driven diversification can be now dealt with refined methods (Ives et al., 2007; FitzJohn 2010; FitzJohn et al., 2014), but low phylogenetic signals seem more difficult to deal with because they always worsen trait prediction. Unless the new statistical models significantly improve the accuracy of PTP at low phylogenetic signals (Guénard et al., 2013; Elliot and Mooers, 2014; Revell, 2014), other methods should be used instead.
Recent methods claim for the use of genomic information to infer microbial traits that are hard to measure (Lauro et al., 2009; Barberán et al., 2014; Fierer et al., 2014). This method confidently assigns traits to those sequenced microbes that are closely related to representatives of sequenced genomes. But how close is closely related? Again, the answer depends on the phylogenetic conservatism of the trait. The lower the phylogenetic signal, the closer the sequenced representative should be. Barberán et al. (2014) assigned genomic traits to operational taxonomic units whose 16S rRNA sequence differed in ⩽1% of the bases to the query genome (that is, at ⩾99% sequence identity). Such a stringent cutoff allowed assigning traits to only 500 out of a total 124 000 sequenced operational taxonomic units. This brute force approach will increase its efficiency as more genomes become sequenced. But, at the same time we massively sequence genomes we also need to culture microbes and record traits of ecological relevance that cannot be inferred from genomes. The scarcity of information on direct measures of microbial phenotypes has slowed down the development of trait-based approaches in the study of microbial communities. Altogether, these findings should prompt scientists to continue phenotyping microbes as taxonomists have always done to characterize the diagnostic characters. This is the way our phylogenetic signal database may grow in the future and, together with the PTP guidelines provided here, facilitate ecologists to infer microbial traits easily.
Acknowledgments
We thank Helen Morlon for inspiring this article based on her comments during the review process of a previous article and Juanjuan Chai, Chongle Pan and co-authors for kindly providing us with the necessary information on their study to calculate phylogenetic signals. We also thank Santiago Donat for technical assistance during the compilation of phylogenetic signals from the literature, and several anonymous referees for their thoughtful comments on the manuscript. Financial support was provided by the Spanish Ministry of Science and Innovation (R+D Projects CGL2011-29585-C02-01 and CGL2014-58333-P). MG acknowledges support by the Ramón y Cajal Programme (Spanish Ministry of Economy and Competitiveness). Calculations were partly performed in the Supercomputing Centre of Galicia (CESGA).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Angly FE, Dennis PG, Skarshewski A, Vanwonterghem I, Hugenholtz P, Tyson GW. (2014). Copyrighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Ramirez KS, Leff JW, Bradford MA, Wall DH, Fierer N. (2014). Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol Lett 17: 794–802. [DOI] [PubMed] [Google Scholar]
- Blomberg SP, Garland T Jr, Ives AR. (2003). Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57: 717–745. [DOI] [PubMed] [Google Scholar]
- Brown MV, Ostrowski M, Grzymski JJ, Lauro FM. (2014). A trait based perspective on the biogeography of common and abundant marine bacterioplankton clades. Mar Genom 15: 17–28. [DOI] [PubMed] [Google Scholar]
- Bruins MR, Kapil S, Oehme FW. (2000). Microbial resistance to metals in the environment. Ecotox Environ Safe 45: 198–207. [DOI] [PubMed] [Google Scholar]
- Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. (2011). Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA 108: 14288–14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IG, Kim SH. (2007). Global extent of horizontal gene transfer. Proc Natl Acad Sci USA 104: 4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W. (2008). Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci USA 105: 10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot MG, Mooers AØ. (2014). Inferring ancestral states without assuming neutrality or gradualism using a stable model of continuous character evolution. BMC Evol Biol 14: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan WF, Pearson YE, Larsen EA, Lynch HJ, Turner JB, Staver H et al. (2013). Phylogenetic prediction of the maximum per capita rate of population growth. Proc R Soc Lond B 280: 20130523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Barberán A, Laughlin DC. (2014). Seeing the forest for the genes: using metagenomics to infer the aggregated traits of microbial communities. Front Microbiol 5: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzJohn RG, Pennell MW, Zanne AE, Stevens PF, Tank DC, Cornwell WK. (2014). How much of the world is woody? J Ecol 102: 1266–1272. [Google Scholar]
- FitzJohn RG. (2010). Quantitative traits and diversification. Syst Biol 59: 619–633. [DOI] [PubMed] [Google Scholar]
- Fraser C, Hanage WP, Spratt BG. (2007). Recombination and the nature of bacterial speciation. Science 315: 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freckleton RP, Harvey PH, Pagel M. (2002). Phylogenetic analysis and comparative data: s test and review of evidence. Am Nat 160: 712–724. [DOI] [PubMed] [Google Scholar]
- Fritz SA, Purvis A. (2010). Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits. Conserv Biol 24: 1042–1051. [DOI] [PubMed] [Google Scholar]
- Garland T Jr, Ives AR. (2000). Using the past to predict the present: Confidence intervals for regression equations in phylogenetic comparative methods. Am Nat 155: 346–364. [DOI] [PubMed] [Google Scholar]
- Goberna M, Navarro JA, Valiente-Banuet A, García C, Verdú M. (2014). Abiotic stress tolerance and competition related traits underlie phylogenetic clustering in soil bacterial communities. Ecol Lett 17: 1191–1201. [DOI] [PubMed] [Google Scholar]
- Green JL, Bohannan BJM, Whitaker RJ. (2008). Microbial biogeography: from taxonomy to traits. Science 320: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Guénard G, Legendre P, Peres-Neto P. (2013). Phylogenetic eigenvector maps: a framework to model and predict species traits. Methods Ecol Evol 4: 1120–1131. [Google Scholar]
- Ives AR, Midford PE, Garland T Jr. (2007). Within-species measurement error in phylogenetic comparative methods. Syst Biol 56: 252–270. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Wu M, Eisen JA, Green JL. (2012). Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLOS Comput Biol 8: e1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Canback B, Berg OG. (2003). Horizontal gene transfer: A critical review. Proc Natl Acad Sci USA 100: 9658–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotech 31: 814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S et al. (2009). The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA 106: 15527–15533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorén JG, Farfán M, Fusté MC. (2014). Molecular phylogenetics and temporal diversification in the genus Aeromonas based on the sequences of five housekeeping genes. PLoS One 9: e88805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins EP, Hansen TF. (1997). Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am Nat 149: 646–667. [Google Scholar]
- Martiny AC, Treseder K, Pusch G. (2013). Phylogenetic conservatism of functional traits in microorganisms. ISME J 7: 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme D, Freckleton R, Thomas G, Petzoldt T, Fritz S, Nick I et al. (2013). Caper: comparative analyses of phylogenetics and evolution in R. R package version 0.4. Available from http://CRAN.R-project.org/package=caper.
- Pagel M. (1999). Inferring the historical patterns of biological evolution. Nature 401: 877–884. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Letunic I, Yamada T, Jensen LJ, Bork P. (2011). Toward molecular trait-based ecology through integration of biogeochemical, geographical and metagenomic data. Mol Syst Biol 7: 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ, Harmon LJ, Collar DC. (2008). Phylogenetic signal, evolutionary process, and rate. Syst Biol 57: 591–601. [DOI] [PubMed] [Google Scholar]
- Revell LJ. (2012). phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3: 217–223. [Google Scholar]
- Revell LJ. (2014). Ancestral character estimation under the threshold model from quantitative genetics. Evolution 68: 743–759. [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E et al. (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA 110: 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira-Silva S, Rocha EPC. (2010). The systemic imprint of growth and its uses in ecological (meta)genomics. PLOS Genet 6: e1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN et al. (2009). A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462: 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneveld JRR, Thurber RLV. (2014). Hidden state prediction: a modification of classic ancestral state reconstruction algorithms helps unravel complex symbioses. Front Microbiol 5: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.