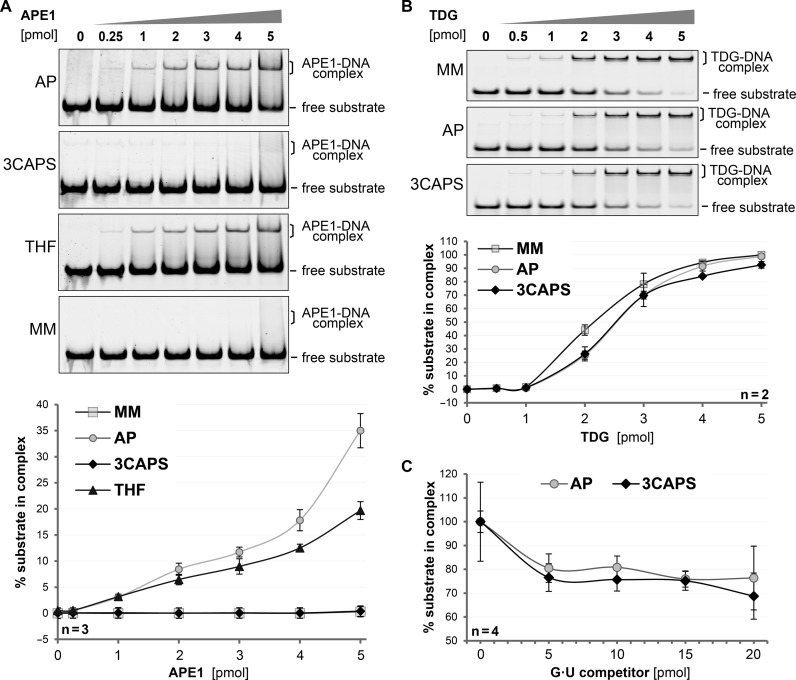
Figure 3.
The structural integrity of the synthetic AP–site analogue 3CAPS. DNA substrates containing either G•U mismatches (MM), enzymatically produced AP-sites (AP) or the synthetic analogues 3CAPS and THF were incubated with increasing amounts of APE1 or TDG. The formation of DNA–protein complexes was analysed by EMSA. (A) Binding of APE1 to 250 fmol of substrate DNA was quantitatively analysed. (B) Binding of TDG to 1 pmol of labelled DNA substrate was quantitatively analysed in the presence of non-labelled homoduplex DNA to block unspecific binding. (C) For competition experiments, 1 pmol of 3CAPS or natural AP–site substrate was incubated with 4 pmol of TDG prior to the addition of the indicated amounts of G•U competitor heteroduplex DNA. Error bars, SEM of the indicated number of experiments.