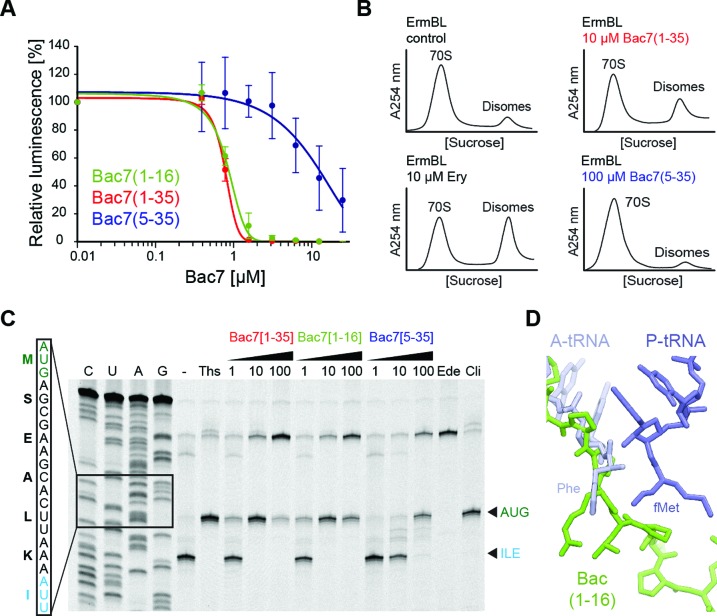
Figure 4.
Mechanism of action of Bac7 on the ribosome. (A) Effects of increasing concentrations of Bac7 derivatives Bac7(1–16) (green), Bac7(1–35) (red) and Bac7(5–35) (blue) on the luminescence resulting from the in vitro translation of firefly luciferase (Fluc) using an Escherichia coli lysate-based system. The error bars represent the standard deviation from the mean for triplicate experiments and the luminescence is normalized relative to that measured in the absence of peptide, which was assigned as 100%. (B) Sucrose gradient profiles to monitor disome formation from in vitro translation of the 2XErmBL mRNA in the absence (control) or presence of 20 μM erythromycin (Ery), 10 μM Bac7(1–35) (red) or 100 μM Bac7(5–35) (blue). (C) Toe-printing assay performed in the absence (−) or presence of increasing concentrations (1, 10, 100 μM) of Bac7(1–35), Bac7(1–16) or Bac7(5–35), or 100 μM thiostrepton (Ths), 50 μM edeine (Ede) or 50 μM clindamycin (Cli). Sequencing lanes for C, U, A and G and the sequence surrounding the toe-print bands (arrowed) when ribosomes accumulate at the AUG start codon (green, initiation complex) or the isoleucine codon (blue, stalled elongation complex) are included for reference. (D) Structural comparison of Phe-tRNAPhe (slate) in the A-site and fMet-tRNAiMet in the P-site (blue) (26) with the binding site of Bac7(1–16) (green).