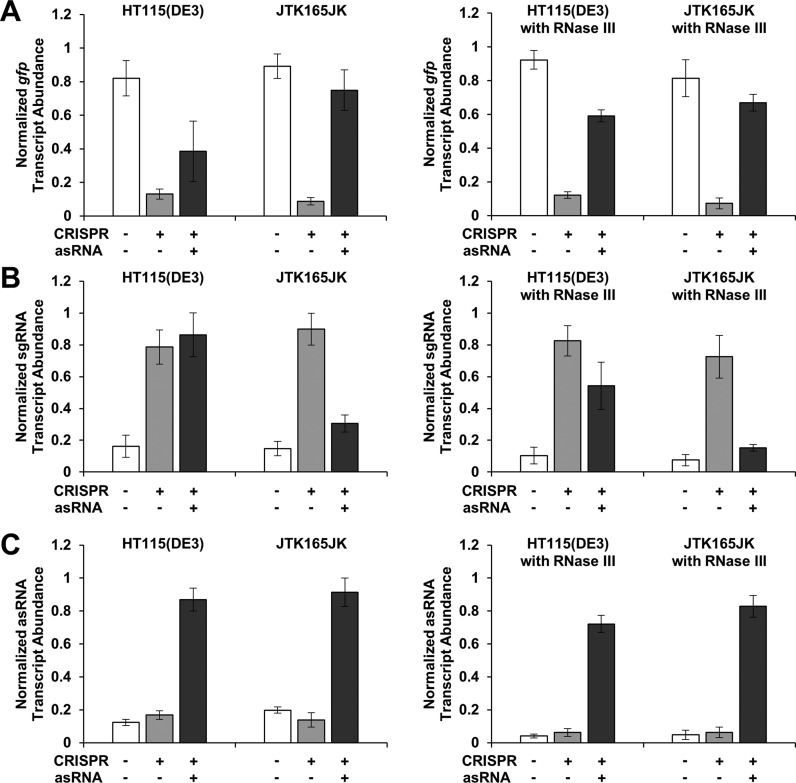
Figure 5.
Derepression is affected by RNase III. Normalized transcript abundance of gfp (A), sgRNA (B) and asRNA (C) in HT115(DE3), JTK165JK and two rescue strains (with a plasmid containing the RNase III gene; right figures ‘with RNase III’) is shown, as measured by RT-qPCR. HT115(DE3) is an RNase III mutant strain (43) while JTK165JK has a functional RNase III (42). The rescue strains are HT115(DE3) and JTK165JK with RNase III heterologously expressed from a low-copy number plasmid (51). For better comparison, the original strains HT115(DE3) and JTK165JK were also transformed using the same backbone plasmid that does not contain the RNase III gene (left figures). RNase III binds to and degrades double-stranded RNA (49,50), including the sgRNA–asRNA complex. The measured relative transcript abundance of gfp, sgRNA (sgR14-T6) and asRNA (asRS6) were divided by that of three reference genes (cysG, hcaT and idnT) and normalized to the relative transcript abundance of the corresponding positive control. The positive controls are cells with either GFP-, sgRNA-, or asRNA-containing plasmid. GFP is under the control of the constitutive Bba J23104 promoter. Cells were grown in the presence of 0.2 ng/ml aTc and 0.25 mM IPTG (+ for CRISPR); and for asRNA, without (−) or with (+) 5 mM Ara (see Figure 1C for the schematic). See Supplementary Figure S5 for the fluorescence results obtained from HT115(DE3), JTK165JK and two rescue strains. The error bars represent the standard deviation of the normalized transcript abundance values from three biological replicates (two technical replicates each; total six replicates) performed on three different days.