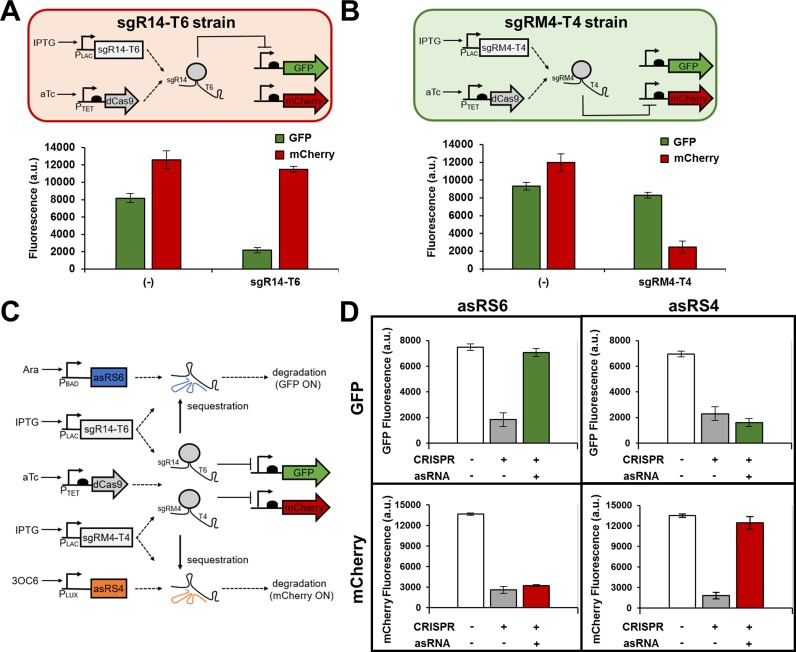
Figure 6.
Orthogonal control of gene expression. Genetic circuits showing the target specificity of sgR14-T6 (A) and sgRM4-T4 (B) in the presence of two reporter genes, gfp and mCherry. Each sgRNA was tested independently in JTK165JK (sgR14-T6 and sgRM4-T4 strains) and specifically represses its cognate gene, but not the other gene. The induced cells were grown in the presence of 0.2 ng/ml aTc and 0.25 mM IPTG. (−) indicates that either sgR14-T6 strain or sgRM4-T4 strain was grown with no inducer. (C) A genetic circuit showing an orthogonal control of GFP and mCherry through the combined CRISPR and asRNA system. The sgR14-T6 represses GFP, and sgRM4-T4 represses mCherry as shown in (A) and (B). The asRS6 binds to T6 to derepress GFP, and the asRS4 binds to T4 to derepress mCherry. Both sgR14-T6 and sgRM4-T4 are simultaneously transcribed by the IPTG-inducible PLAC promoter. (D) Multiple sgRNAs and their cognate asRNAs can specifically repress or derepress GFP and mCherry in the same cell. Samples were grown in the presence of 1 ng/ml aTc and 0.5 mM IPTG (transcribes both sgR14-T6 and sgRM4-T4) to achieve simultaneous repression of GFP and mCherry (+ for CRISPR). The derepression of either GFP or mCherry was achieved by growing the cells in the presence of 1 ng/ml aTc and 0.5 mM IPTG, and either 5 mM Ara (for GFP) or 5 mM 3OC6 (for mCherry). GFP and mCherry are under the control of the constitutive Bba J23104 and Bba J23100 promoters, respectively. The fluorescence (a.u.) was measured using a microplate reader. The error bars represent the standard deviation of the fluorescence values from three biological replicates performed on three different days.