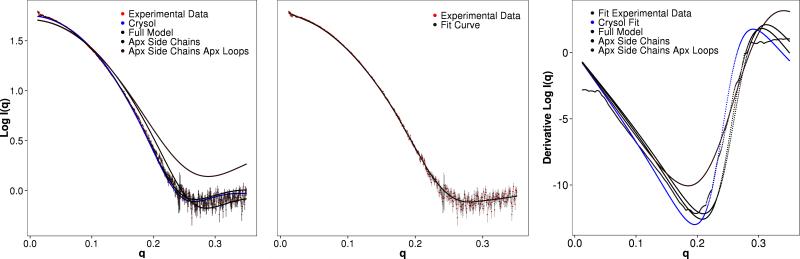
Figure 4. Depiction of the Experimental SAXS profile for Hen Egg White Lysozyme and SAXS profiles computed with BCL::SAXS for different protein states.
Panel A represents the fit on a log10 scale with Experimental data being the SAXS profile or Hen Egg White lysozyme, Crysol is the curve generated through Crysol from 6lyz and fit to the experimental data. Full Model is the curve generated through BCL::SAXS from 6lyz. Apx Side Chains is the curve generated through BCL::SAXS using Backbone atoms only and summing the form factors for all side chain atoms at the Cβ coordinate of the residue. Apx Side Chains Apx Loops is the curve generated through BCL::SAXS using loop approximation and side chain approximation. Panel B shows the locally weighted scatterplot smoothing (LOESS) of the experimental SAXS data points. Panel C shows the fit of previous data types from panel A using the derivative of the log10 profiles.