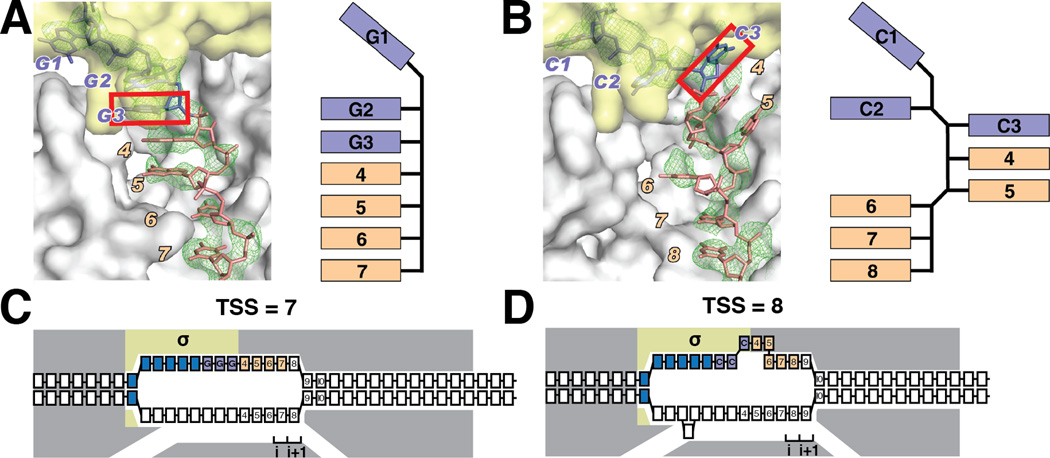
Fig. 4. Crystal structures define paths of DNA nontemplate-strands with representative RRR and YYY discriminators.
(A) Crystal structure of an initiation complex with RRR discriminator and nontemplate-strand length corresponding to TSS at position 7 (RPo-GGG-7). Left, simulated annealing |Fo-Fc| omit map contoured at 2.3σ and atomic model for interactions of RNAP with DNA nontemplate-strand. Right, schematic path of DNA. Gray, RNAP; yellow, σ; purple, discriminator nucleotides; pink, nontemplate-strand nucleotides downstream of discriminator; green, simulated annealing |Fo-Fc| omit map contoured at 2.3σ. Red box, third nucleotide of discriminator.
(B) Crystal structure of an initiation complex with YYY discriminator and nontemplate-strand length corresponding to TSS at position 8 (RPo-CCC-8; symbols as in A).
(C–D) Interpretation: The different position of the third nucleotide of the discriminator in the structure with a YYY discriminator increases the distance between the third nucleotide of the discriminator and downstream duplex DNA, accommodating an additional nucleotide in the connector.